Medicine Group . 2023 January 28;4(1):077-083. doi: 10.37871/jbres1650.
Molecular Diversity and Characteristics of KRAS Mutations in Colorectal Cancers
Amrit Kaur Kaler1*, Omkar Dharut1, Smitha Umarji1, Shweta Limaye1, Bijal Kulkarni2, Meenal Hastak2, Nivetha Athikari2, Yash Tiwarekar1, Samrudhi Rane1, Ankita Nikam1, Imran Sheikh3 and Sandeep Goyle3
2Department of Pathology, Kokilaben Dhirubhai Ambani Hospitals & Medical Research Institute, Mumbai, India
3Department of Oncology, Kokilaben Dhirubhai Ambani Hospitals & Medical Research Institute, Mumbai, India
- KRAS
- Colorectal cancers
- Codons
- Molecular diversity
- Targeted treatment
- Targeted treatment
- Left side
- Epidemiology
Abstract
Background: KRAS oncogene is involved in colorectal carcinogenesis in 22 to 45% of cases. KRAS mutations can render Epidermal Growth Factor Receptor (EGFR) inhibitors ineffective. The present study is aimed to determine the molecular diversity, frequency, and characteristics of KRAS mutations in metastatic Colorectal Cancer (mCRC). The gene KRAS contains 4 coding exons and 1 non-coding exon, of which exon 2 has the highest mutation rate, which is directly associated with the occurrence of poor prognosis and drug resistance. Codon 12, 13, 61 are common KRAS mutations. The most prevalent KRAS mutation observed was codon 12-G12D mutation where Glycine is changed to Aspartic acid.
Materials and Methods: A retrospective study was conducted on both in-situ and metastatic colorectal cancers that were registered for KRAS gene mutation testing in Department of Molecular Pathology and Genomics, Kokilaben Dhirubhai Ambani Hospital & Medical Research Institute, from January 2016 to January 2022. KRAS mutations studies were carried out in primary and metastatic 87 patients of colorectal cancers. The samples contained 37 women aged 25-80 years and 50 men aged 29-77 years. Formalin fixed and paraffin embedded tissue samples were evaluated using Sanger sequencing and statistical analysis was carried out.
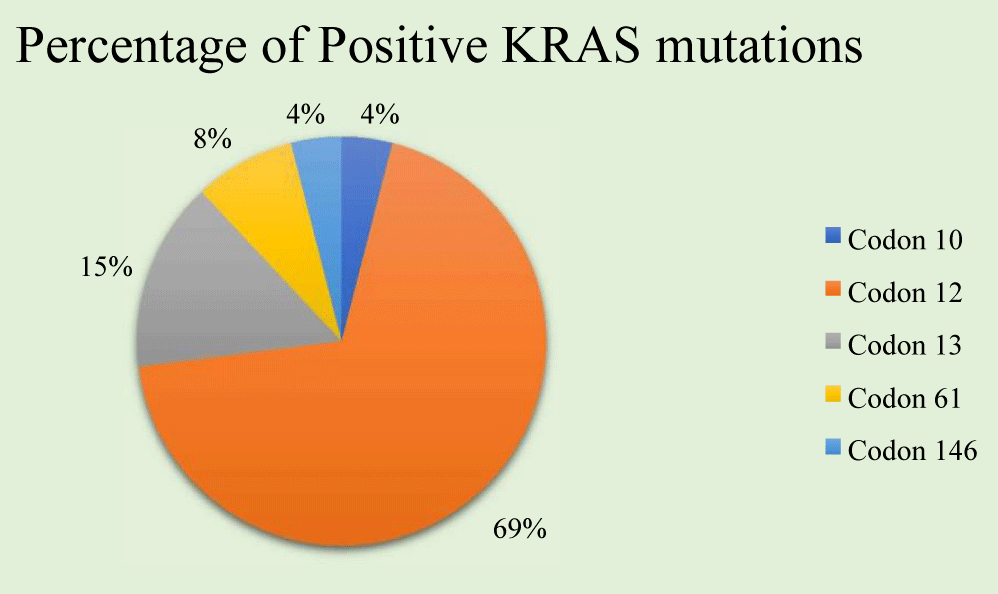
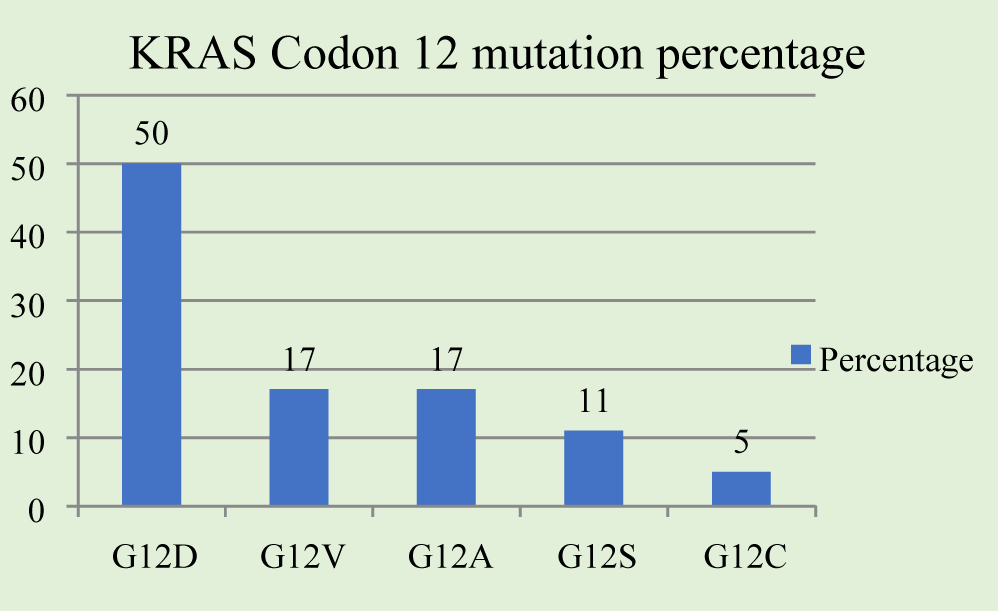
Results: KRAS mutations were demonstrated in 26 cases of 87 patients (29.88%). Of the 26 cases, the substitution nucleotide variations as point mutations were as follows: Codon 12-18 cases (69%), codon 13-4 cases (15%), codon 61-2 cases (8%), codon 146-1 case (4%), and codon 10-1 case (4%). The Codon 12 mutations were detected as follows: G12D-9 cases (50%), G12V-3 cases (17%), G12A-3 cases (17%), G12S-2 cases (11%), G12C-1 case (5%); Codon 13 showed all 4 mutations (4%) in G13D. KRAS mutations were found to be more frequent in elderly age group. Most m-KRAS tumors were located in sigmoid colon and rectum (left sided/distal colon), although, the mutations were spread across including ascending colon (right sided/proximal colon) and rarely in jejunum too. The TNM stage showed majority (50%) of KRAS mutations in stage IV, and 20% of cases in stage II and III each.
Conclusion: In this series, 29% of colorectal cancer tissue samples had KRAS mutations. Their frequency and distribution showed majority mutations on left sided/distal colon, elderly population, more common in females and presented at an advanced stage. There was no clinicopathological significance to histomorphology. Understanding KRAS mutations in different aspects can give a better comprehensive understanding of KRAS associated CRCs.
Abbreviations
KRAS: Kirsten Rat Sarcoma viral oncogene; EGFR: Epidermal Growth Factor Receptor; mCRC: metastatic Colorectal Cancer; TNM stage: T: Size of the tumor and any spread of cancer into nearby tissue, N: Spread of cancer to nearby lymph nodes, M: Metastasis (spread of cancer to other parts of the body); FDA: Food and Drug Administration; MSS: Microsatellite Stable; PCR: Polymerase Chain Reaction; FFPE: Formalin-Fixed Paraffin-Embedded; GTPase: Guanine Tri Phosphatase
Introduction
Colorectal Cancer (CRC) is a third most common worldwide and usually presents at an advanced stage and shows high mortality [1]. Recent advances in molecular targeted therapeutic approaches to CRC has led to the approval of Cetuximab and Panitumumab, Antiepidermal Growth Factor Receptor (EGFR) targeted therapies by US Food and Drug Administration (FDA) in 2009 [2,3]. Kirsten Rat Sarcoma viral oncogene homolog (KRAS) gene is approximately seen 35.0-45.0% of the CRC cases [4]. KRAS is known to be a poor prognostic marker in CRC, which is still debatable based on the type of mutation and location [5]. KRAS is found be usually seen in Microsatellite Stable Tumors (MSS) which is responsive to the 5Fluoro-Uracil in stage II-III cancers [6,7]. Three distinct molecular pathways have been defined in CRC: Microsatellite Instable (MSI) phenotypic tumors; Chromosomal Instability (CIN) phenotype; CpG Island Methylator Phenotype (CIMP) [8]. The significance of the Cancer Genome Atlas (TCGA) integrated molecular classification have been clearly laid out. [9] KRAS is known to be associated with CIN molecular pathway and is also biological marker of resistance to targeted therapy [10].
Somatic KRAS mutations in codons 12, 13 (exon 2), 59, 61 (exon 3), and 146 (exon 4) are common hotspots. Other non-hotspot mutations in codons 2, 3, 4, 63 and 154 occur less frequently in CRC. Most of the reported mutations are single nucleotide point mutations, particularly G > A transitions and G > T transversions [11]. According to published western study by Tan C, among the codon 12 mutations, p.Gly12Asp, and p.Gly12Val are the most frequent, while in codon 13, the substitution of Glycine by Aspartate (p.Gly13Asp) is the most common [3]
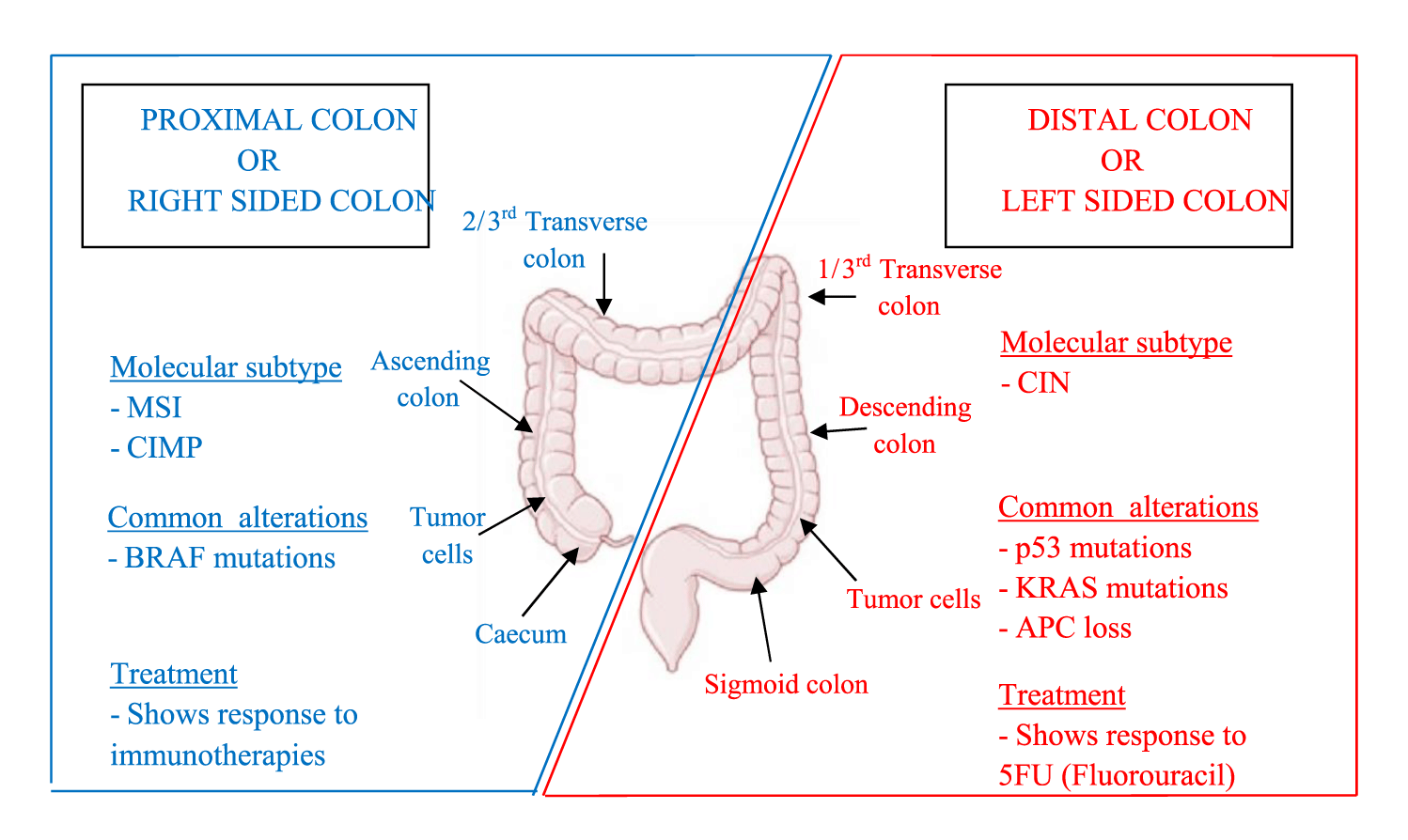
KRAS has been found to be more common on the left sided location along with other genes like APC, PIK3CA, p53 mutations [5]. The mutation usually results in constitutionally active RAS protein, leading to cell proliferation and cell survival. It is essential to investigate the underlying mutations in the KRAS gene in mCRC patients to identify the prevailing patterns based on topographical location of the tumor i.e. either right sided/proximal colon or left sided/distal colon (Figure 1) [12]. Two clinical trials have shown that KRAS-Wild (WT) left sided CRC have survived longer when treated with Cetuximab; whereas KRAS-WT right sided CRC showed longer survival when Bevacizumab was added to the treatment regimen.
(ClinicalTrials.gov identifier: NCT00265850) [13]. In metastatic settings, KRAS-Mutant (MT) patients showed worse overall survival than KRAS-WT type patients [14].
KRAS has been difficult gene to target due to a unique intrinsic structure of the KRAS protein, the surface of which is a smooth and shallow leading to difficulty in binding of drugs. However, Sotorasib and Onvansetinib, new targetable treatments against KRAS p.12C are able to bind GTP binding pocket and has been approved by FDA [15,16].
Materials and Methods
A retrospective descriptive study was conducted on both in-situ and metastatic colorectal cancers (n = 87) that were registered for KRAS gene mutation testing in the Department of Molecular Pathology and Genomics from January 2016 to January 2022 at Kokilaben Dhirubhai Ambani Hospital & Medical Research Institute, Mumbai. Using patients’ medical records, data was collected regarding variables such as demographic characteristics (age and gender) and pathological tumor features (primary site, and staging).
Around 10 Curls from the patient's Formalin-Fixed Paraffin-Embedded (FFPE) tissue blocks were obtained and carried out DNA isolation by QIAamp DNA FFPE Tissue Kit (Qiagen, Hilden, Germany). The slides were specifically chosen by Molecular Pathologist to include tumor tissue with > 30% tumor cells without significant necrosis, mucin, or inflammation [17]. All the tumor sections were placed in two baths of xylene for deparaffinization and after, for rehydratation, in two baths of ethanol. For a complete lysis, they were placed in the lysis buffer with proteinase K at 56°C overnight. After lysis, DNA was precipitated with ethanol and was fixated on the QIAamp silica membrane by centrifugation. Two washes were performed and the purified DNA was eluted in buffer AE or water. The purified DNA was used for Polymerase Chain Reaction (PCR). Results of the PCR amplification reaction were visualized using gel electrophoresis. The products were subjected to post amplification purification followed by Sanger's sequencing by Capillary electrophoresis on ABI 3500Dx Genetic Analyzer. Data were analyzed with respect to age groups, gender, KRAS genotype, location on right/left side and TNM classification using standard descriptive statistics [15].
Results
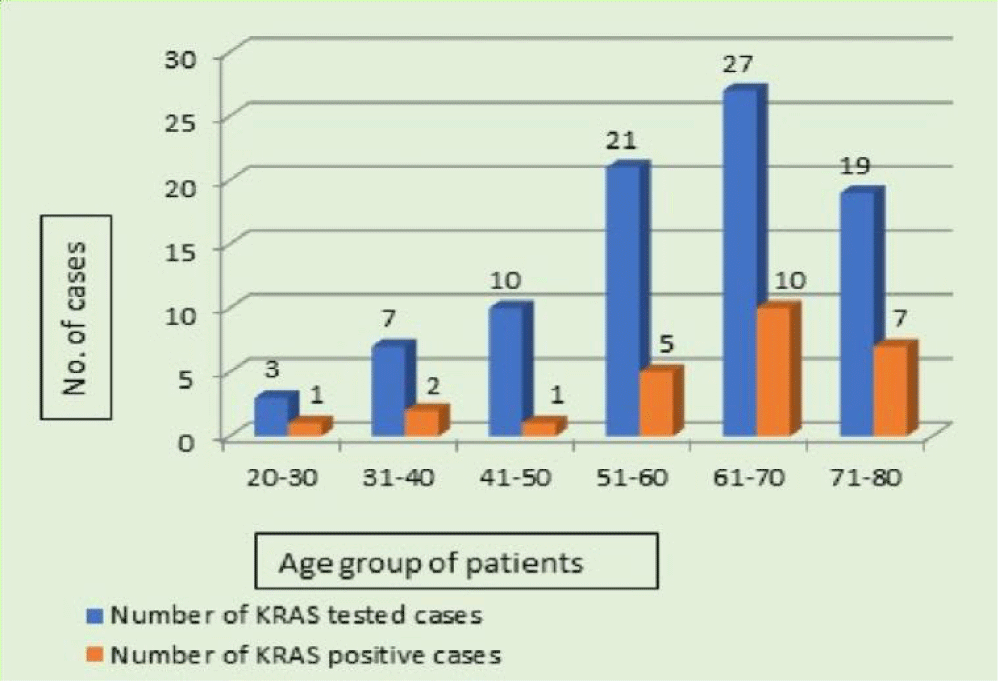
The colorectal tissue samples of 87 patients were tested with age between 20 to 80 years (median age, 63 years). Of these, 26 (30%) tested positive for KRAS mutations. The remaining 61 (70%) samples had the wild-type allele. Overall, there were 50 (57%) males and 37 (43%) females. Among the KRAS positive cases, there were 12 (45%) males and 14 (54%) females. The highest percentage of mutant KRAS cases was found in the 50-70 years age group (Figure 2).
Out of these 26 positive cases, the main point mutations were as follows: codon 12-18 (69%), codon 13-4 (15%), codon 61-2 (8%), codon 146-1(4%), and codon 10-1(4%) (Figure 3).
The codon 12 mutations showed the following mutations: G12D-9 (50%), G12V-3 (17%), G12A-3 (17%), G12S-2 (11%), G12C-1 (5%), (Figure 2) and codon 13 mutation observed was G13D-4. The p.Gly12Asp (c.35G > A) mutation had the highest frequency in both females -7 (27%) and males -2 (8%). Multiple mutations in the same individual were not detected in any of the patients in this cohort.
Discussion
KRAS is located on Chromosome 12p12.1, is a member of RAS family of genes and it is responsible for oncogenesis in various malignancies and the sustained signalling makes it vulnerable to high profile therapeutic target [19]. Activated KRAS binds to GTPase protein, produces a conformational change in the KRAS protein which affects its interaction with the downstream transducer GTPase Activating Proteins (GAPs) [19]. KRAS is the one of the most common gene mutations occurring in 17-25% of all cancers [20]. Approximately, 30-40% of colorectal cancer carries KRAS mutation which are associated with poor prognosis and advanced disease [21,22]. The overall frequency of positive KRAS mutations observed in this study was 30% which is slightly lesser than the reported studies. There are significant differences between the frequencies of KRAS gene mutation in patients with CRC from different continents: Asia -24%, Europe -36% and Latin America - 40% as reported by Yamane L, et al. [23].
The present study cohort is widely spread across age groups from 21 to 80 years, with most mutations were found in age group 61-70 (38%) followed by 71-80 (27%) and 51 to 60 (19%) and less commonly in younger patients (Figure 4). An Indian study by Veldore, et al. [24] showed similar findings with respect to age. Conversely, Ozer M, et al. [25] in their study cohort reported 42% KRAS mutations in of young patients, which is higher than previously reported studies. They also inferred that prognosis was better in patients with less than 50 years, and in between 50-70 years. The prognostic impact is lost above 70 years due to underutilization of targeted therapy and more vulnerability to chemotherapy. Further studies might elucidate the clinical characteristics of young-onset colorectal cancer. The prevalence of KRAS positive mutations was higher in females (54%) as compared to males (45%) in the present study. This was contrary to the Srilankan study by Sirisena ND, et al. [26] where they reported KRAS mutations in 56% of males and 44% in females.
The distribution of mutations in different codons in KRAS was studied and the majority of KRAS mutation are mainly present in codon 12, 13 and codon 64 together corresponding to 92% positivity. The mutations on minor (146, 10) codons were concluded to 8%. Knijn N, et al. [27] reported 95% KRAS mutations in codon 12 and 13; other mutations in codons 61, 146 in a rare scenario, accounting for 5% of all the mutations. Specific mutations at different codons are known modify the biologic behaviour of the tumor. KRAS mutational subtypes in the present study did not show any differentiation into mucinous/nonmucinous morphology. Further studies will help to understand further associations with morphogenomics.
In the study, the most common mutation detected on codon 12 was G12D (c.35G > A; p.Gly12Asp) which accounted for 50% of the mutations; followed by 17% each of G12V (c.35G > T; p.Gly12Val) and G12A (c.34G > A; p.Gly12Ser) mutations. The most common KRAS mutation types observed are p.Gly12Asp (Codon 12), p.Gly12Val (Codon 12)and p.Gly13Asp (Codon 13), accounting for almost 62% of all mutations. Overall studies have shown that these three mutations account for around 70% of the codon 12 mutations [28,29].
All 26 cases of KRAS were located to different anatomical sites and vastly grouped into left sided/distal and right sided/proximal location. The tumor site with higher incidence is left sided colon composed of 1/3 transverse colon, descending colon, sigmoid colon and rectum constituting 58% of total mutations. G12D was found to be the most common mutation (33%) on left side colon. This was similar to the study reported by Indian Study, Turkish and Chinese Study [24,30,31]. But Watanabe, et al. [32] found a higher frequency of KRAS mutation in right sided colon cancers rather than left sided colonic cancers. In an interesting finding, 1 case of codon 13 (G13D) mutations was located on jejunum which is a rare finding.
The clinical correlation also elucidated that KRAS mutations were found in late stage of presentation of the colorectal cancer. The results showed 20% of cases in stage 2, 20% in stage III, and 50% in stage IV (Table 1). Inoue, et al. [2] also reported similar findings with the frequency of KRAS mutation was 20% in stage I, 30% in stage II, 40% in stage III and 58% in stage IV.
| Table 1: Distribution of KRAS mutation depending on location and stage. | ||||||||||
| KRAS Codon 10 |
KRAS Codon 12 | KRAS Codon 13 |
KRAS Codon 61 |
KRAS Codon 146 |
||||||
| G12D | G12V | G12C | G12S | G12A | ||||||
| Gender (n = 26) | Male = 12 | 0 | 2 | 3 | 0 | 2 | 1 | 2 | 2 | 0 |
| Female = 14 | 1 | 7 | 0 | 1 | 0 | 2 | 2 | 0 | 1 | |
| Left Sided Colon (n = 18/26) |
Descending colon, Sigmoid colon, Rectum, 1/3 transverse colon |
1 | 6 | 3 | 1 | 2 | 3 | 2 | 0 | 0 |
| Right Sided Colon (n = 7/26) |
Caecum, Ascending colon, appendix, 2/3 transverse colon | 0 | 3 | 0 | 0 | 0 | 0 | 1 | 2 | 1 |
| Jejunum (n = 1/26) | Small Intestine | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| Pathologica l Stage (n = 16) | T1 | |||||||||
| T2 | 1 | 1 | 1 | |||||||
| T3 | 1 | 2 | ||||||||
| T4 | 1 | 4 | 1 | 1 | 1 | 2 | ||||
| *p value couldn’t be calculated as the sample size for positive KRAS mutations was less (n = 26). | ||||||||||
Novel Mutation: A rare case of codon 10 insertion was found in our study, also known as codon 10 duplication. Codon 10 mutation is a rare case of insertion where GGA is inserted that leads to two peaks from that codon in the electropherogram. The patient has a metastatic sigmoid cancer. The metastasis has reached to the abdominal skeletal muscles. Patient has been treated with FOLFIRI (Leucovorin + 5FU + Irinotecan) regimen of chemotherapy along with Bevacizumab, an anti-angiogenic immunotherapy drug. This rare mutation 10G11, which suggested the insertion of an additional Glycine residue between Glycine (amino acid 10) and Alanine (amino acid 11). Only 2 cases have been reported of which the first was reported in the patient with myeloid leukemia and the other in colorectal cancer [33,34].
Another rare mutation of codon 146 was found on the right sided colon and required further investigation.
KRAS is a negative prognostic factor with the mutational diversity is potentially oncogenic by conveying selective growth advantage to the cells and potentially contributes to primary resistance for anti-EGFR mAb targeted therapy [2,10]. The KRAS mutation can still respond to 5-Fluorouracil (5-FU) based regimens but doesn’t predict response to 5-FU [5]. Historically, KRAS has been known to be untargetable, probably due to high affinity to cytoplasmic GTP, RAS protein with no pocket or groove for binding the drug and multiple resistance mechanism due to activation of bypass upstream and downstream signals. [35] Sotorasib, a small molecule inhibitor, is the first KRAS targeting drug approved by FDA in KRASp.G12C positive lung cancers which covalently binds novel surface groove and locks the KRAS 12C protein irreversibly [15][36] Onvansertinib, a pole like kinase inhibitor (PLK-1) has been approved with KRAS mutant metastatic CRC in combination with FOLFIRI (5-Fluorocil, Leucovorin, and Irinotecan) and Bevacizumab. [16] The resistance mechanism include reactivaton of receptor tyrosine kinase (RTK) pathway by an alternate mechanism, where the combination therapy of targeted pathways with drugs targeting RTKs can be a promising strategy.
Adagrasib, a new drug approved in United States in December 2022, developed for treating non-small cell lung cancer for KRAS G12C mutated patients, can be a promising discovery also in relation with CRC after further research and development [37].These characteristics of the study cohort can help pathologists and oncologists to thoroughly understand the behaviour of mutations and will increase efficiency in the approach towards managing the disease.
Conclusion
In the present study, 29% of colorectal cancer tissue samples were positive for KRAS mutations. KRAS 12D was found to be the most common mutational site. The majority of mutations were detected on left sided/distal colon, in elderly population, more common in females and presented at an advanced TNM stage. Clinicopathological significance due to histomorphology was not observed in the given study.
Using KRAS testing to restrict the use of EGFR inhibitor therapy would help to select the appropriate patients. Understanding drug development through binding of development drugs at different mutational sites, might give a comprehensive understanding of KRAS inhibition. Hence, KRAS is a promising biomarkers and shows both prognostic and predictive value.
Acknowledgment
We express our gratitude towards Mrs. Varsha Vadera for always supporting us.
Author contributions
Conceptualization: Amrit Kaler; Methodology: Omkar D; Validation: Smita K; Formal analysis: Sweta L; Investigation: Yash T; Data curation: Samrudhi, Ankita; Writing-original draft preparation: Amrit Kaler; Writing-review and editing: Bijal K, Meenal H, Nivetha A; Supervision: Sandeep G, Imran Shaikh; Project administration: Rajesh Mistry; Funding Acquisition, Not applicable; All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki and approved by the Institutional Review Board (or Ethics Committee) of Kokilaben Dhirubhai Ambani Hospital & Medical Research Institute (Protocol code: KDAH/PUB/2022/38) and date of approval: 7 September 2022).
References
- Patil PS, Saklani A, Gambhire P, Mehta S, Engineer R, De'Souza A, Chopra S, Bal M. Colorectal Cancer in India: An Audit from a Tertiary Center in a Low Prevalence Area. Indian J Surg Oncol. 2017 Dec;8(4):484-490. doi: 10.1007/s13193-017-0655-0. Epub 2017 Apr 22. PMID: 29203978; PMCID: PMC5705504.
- Inoue Y, Saigusa S, Iwata T, Okugawa Y, Toiyama Y, Tanaka K, Uchida K, Mohri Y, Kusunoki M. The prognostic value of KRAS mutations in patients with colorectal cancer. Oncol Rep. 2012 Nov;28(5):1579-84. doi: 10.3892/or.2012.1974. Epub 2012 Aug 21. PMID: 22922794.
- Tan C, Du X. KRAS mutation testing in metastatic colorectal cancer. World J Gastroenterol. 2012 Oct 7;18(37):5171-80. doi: 10.3748/wjg.v18.i37.5171. PMID: 23066310; PMCID: PMC3468848.
- Harlé A, Filhine-Tresarrieu P, Husson M, Boidot R, Rouyer M, Dubois C, Leroux A, Merlin JL. Rare RAS Mutations in Metastatic Colorectal Cancer Detected During Routine RAS Genotyping Using Next Generation Sequencing. Target Oncol. 2016 Jun;11(3):363-70. doi: 10.1007/s11523-015-0404-7. PMID: 26661077.
- Baran B, Mert Ozupek N, Yerli Tetik N, Acar E, Bekcioglu O, Baskin Y. Difference Between Left-Sided and Right-Sided Colorectal Cancer: A Focused Review of Literature. Gastroenterology Res. 2018 Aug;11(4):264-273. doi: 10.14740/gr1062w. Epub 2018 Feb 8. PMID: 30116425; PMCID: PMC6089587.
- Etienne-Grimaldi MC, Formento JL, Francoual M, François E, Formento P, Renée N, Laurent-Puig P, Chazal M, Benchimol D, Delpero JR, Letoublon C, Pezet D, Seitz JF, Milano G. K-Ras mutations and treatment outcome in colorectal cancer patients receiving exclusive fluoropyrimidine therapy. Clin Cancer Res. 2008 Aug 1;14(15):4830-5. doi: 10.1158/1078-0432.CCR-07-4906. PMID: 18676755.
- Taieb J, Le Malicot K, Shi Q, Penault-Llorca F, Bouché O, Tabernero J, Mini E, Goldberg RM, Folprecht G, Luc Van Laethem J, Sargent DJ, Alberts SR, Emile JF, Laurent Puig P, Sinicrope FA. Prognostic Value of BRAF and KRAS Mutations in MSI and MSS Stage III Colon Cancer. J Natl Cancer Inst. 2016 Dec 31;109(5):djw272. doi: 10.1093/jnci/djw272. PMID: 28040692; PMCID: PMC6075212.
- Nazemalhosseini Mojarad E, Kuppen PJ, Aghdaei HA, Zali MR. The CpG island methylator phenotype (CIMP) in colorectal cancer. Gastroenterol Hepatol Bed Bench. 2013 Summer;6(3):120-8. PMID: 24834258; PMCID: PMC4017514.
- Müller MF, Ibrahim AE, Arends MJ. Molecular pathological classification of colorectal cancer. Virchows Arch. 2016 Aug;469(2):125-34. doi: 10.1007/s00428-016-1956-3. Epub 2016 Jun 20. PMID: 27325016; PMCID: PMC4978761.
- Yamane L, Scapulatempo-Neto C, Reis RM, Guimarães DP. Serrated pathway in colorectal carcinogenesis. World J Gastroenterol. 2014 Mar 14;20(10):2634-40. doi: 10.3748/wjg.v20.i10.2634. PMID: 24627599; PMCID: PMC3949272.
- Inamura K. Colorectal Cancers: An Update on Their Molecular Pathology. Cancers (Basel). 2018 Jan 20;10(1):26. doi: 10.3390/cancers10010026. PMID: 29361689; PMCID: PMC5789376.
- Charlton ME, Kahl AR, Greenbaum AA, Karlitz JJ, Lin C, Lynch CF, Chen VW. KRAS Testing, Tumor Location, and Survival in Patients With Stage IV Colorectal Cancer: SEER 2010-2013. J Natl Compr Canc Netw. 2017 Dec;15(12):1484-1493. doi: 10.6004/jnccn.2017.7011. PMID: 29223986; PMCID: PMC7458121.
- Heinemann V vWL, Decker T, Kiani A, Vehling-Kaiser U, Al-Batran S. Gender and tumor location as predictors for efficacy: Influence on endpoints in first-line treatment with FOLFIRI in combination with cetuximab or bevacizumab in the AIO KRK 0306 (FIRE3) trial. J Clin Oncol. 2014;32:5s(suppl; abstr 3600).
- Modest DP, Ricard I, Heinemann V, Hegewisch-Becker S, Schmiegel W, Porschen R, Stintzing S, Graeven U, Arnold D, von Weikersthal LF, Giessen-Jung C, Stahler A, Schmoll HJ, Jung A, Kirchner T, Tannapfel A, Reinacher-Schick A. Outcome according to KRAS-, NRAS- and BRAF-mutation as well as KRAS mutation variants: pooled analysis of five randomized trials in metastatic colorectal cancer by the AIO colorectal cancer study group. Ann Oncol. 2016 Sep;27(9):1746-53. doi: 10.1093/annonc/mdw261. Epub 2016 Jun 29. PMID: 27358379; PMCID: PMC4999563.
- Skoulidis F, Li BT, Dy GK, Price TJ, Falchook GS, Wolf J, Italiano A, et al. Sotorasib for Lung Cancers with KRAS p.G12C Mutation. N Engl J Med. 2021 Jun 24;384(25):2371-2381. doi: 10.1056/NEJMoa2103695. Epub 2021 Jun 4. PMID: 34096690; PMCID: PMC9116274.
- Lenz, Heinz-Josef, et al. "A phase 1b/2 trial of the PLK1 inhibitor onvansertib in combination with FOLFIRI-bev in 2L treatment of KRAS-mutated (mKRAS) metastatic colorectal carcinoma (mCRC)." (2022): 100-100.
- Lhermitte B, Egele C, Weingertner N, Ambrosetti D, Dadone B, Kubiniek V, Burel-Vandenbos F, Coyne J, Michiels JF, Chenard MP, Rouleau E, Sabourin JC, Bellocq JP. Adequately defining tumor cell proportion in tissue samples for molecular testing improves interobserver reproducibility of its assessment. Virchows Arch. 2017 Jan;470(1):21-27. doi: 10.1007/s00428-016-2042-6. Epub 2016 Nov 16. PMID: 27853865.
- Sanger Sequencing: Introduction, Principle, and Protocol | CD Genomics Blog. 2022.
- Jancík S, Drábek J, Radzioch D, Hajdúch M. Clinical relevance of KRAS in human cancers. J Biomed Biotechnol. 2010;2010:150960. doi: 10.1155/2010/150960. Epub 2010 Jun 7. PMID: 20617134; PMCID: PMC2896632.
- Anderson MW, Reynolds SH, You M, Maronpot RM. Role of proto-oncogene activation in carcinogenesis. Environ Health Perspect. 1992 Nov;98:13-24. doi: 10.1289/ehp.929813. PMID: 1486840; PMCID: PMC1519627.
- Wu HZ, Xiao JQ, Xiao SS, Cheng Y. KRAS: A Promising Therapeutic Target for Cancer Treatment. Curr Top Med Chem. 2019;19(23):2081-2097. doi: 10.2174/1568026619666190905164144. PMID: 31486755.
- Behl AS, Goddard KA, Flottemesch TJ, Veenstra D, Meenan RT, Lin JS, Maciosek MV. Cost-effectiveness analysis of screening for KRAS and BRAF mutations in metastatic colorectal cancer. J Natl Cancer Inst. 2012 Dec 5;104(23):1785-95. doi: 10.1093/jnci/djs433. Epub 2012 Nov 28. PMID: 23197490; PMCID: PMC3514165.
- Yamane LS, Scapulatempo-Neto C, Alvarenga L, Oliveira CZ, Berardinelli GN, Almodova E, Cunha TR, Fava G, Colaiacovo W, Melani A, Fregnani JH, Reis RM, Guimarães DP. KRAS and BRAF mutations and MSI status in precursor lesions of colorectal cancer detected by colonoscopy. Oncol Rep. 2014 Oct;32(4):1419-26. doi: 10.3892/or.2014.3338. Epub 2014 Jul 18. PMID: 25050586.
- Veldore VH, Rao MR, Prabhudesai SA, Tejaswi R, Kakara S, Pattanayak S, Krishnamoorthy N, Tejaswini BN, Hazarika D, Gangoli A, Rahman SM, Dixit J, Naik R, Diwakar RB, Satheesh CT, Shashidhara HP, Patil S, Gopinath KS, Kumar BS. Prevalence of KRAS mutations in metastatic colorectal cancer: A retrospective observational study from India. Indian J Cancer. 2014 Oct-Dec;51(4):531-7. doi: 10.4103/0019-509X.175371. PMID: 26842186.
- Ozer M, Goksu SY, Sanford NN, Ahn C, Beg MS, Ali Kazmi SM. Age-dependent prognostic value of KRAS mutation in metastatic colorectal cancer. Future Oncol. 2021 Dec;17(35):4883-4893. doi: 10.2217/fon-2021-0650. Epub 2021 Nov 11. PMID: 34758634; PMCID: PMC8890131.
- Sirisena ND, Deen K, Mandawala DEN, Herath P, Dissanayake VHW. The pattern of KRAS mutations in metastatic colorectal cancer: a retrospective audit from Sri Lanka. BMC Res Notes. 2017 Aug 10;10(1):392. doi: 10.1186/s13104-017-2731-5. PMID: 28797274; PMCID: PMC5553606.
- Knijn N, Mekenkamp LJ, Klomp M, Vink-Börger ME, Tol J, Teerenstra S, Meijer JW, Tebar M, Riemersma S, van Krieken JH, Punt CJ, Nagtegaal ID. KRAS mutation analysis: a comparison between primary tumours and matched liver metastases in 305 colorectal cancer patients. Br J Cancer. 2011 Mar 15;104(6):1020-6. doi: 10.1038/bjc.2011.26. Epub 2011 Mar 1. PMID: 21364579; PMCID: PMC3065268.
- He K, Wang Y, Zhong Y, Pan X, Si L, Lu J. KRAS Codon 12 Mutation is Associated with More Aggressive Invasiveness in Synchronous Metastatic Colorectal Cancer (mCRC): Retrospective Research. Onco Targets Ther. 2020 Dec 8;13:12601-12613. doi: 10.2147/OTT.S279312. PMID: 33335401; PMCID: PMC7737549.
- Jones RP, Sutton PA, Evans JP, Clifford R, McAvoy A, Lewis J, Rousseau A, Mountford R, McWhirter D, Malik HZ. Specific mutations in KRAS codon 12 are associated with worse overall survival in patients with advanced and recurrent colorectal cancer. Br J Cancer. 2017 Mar 28;116(7):923-929. doi: 10.1038/bjc.2017.37. Epub 2017 Feb 16. PMID: 28208157; PMCID: PMC5379149.
- Baskin Y, Dagdeviren YK, Calibasi G, Canda AE, Sarioglu S, Ellidokuz H, Oztop I. KRAS mutation profile differences between rectosigmoid localized adenocarcinomas and colon adenocarcinomas. J Gastrointest Oncol. 2014 Aug;5(4):265-9. doi: 10.3978/j.issn.2078-6891.2014.038. PMID: 25083299; PMCID: PMC4110495.
- Xie MZ, Li JL, Cai ZM, Li KZ, Hu BL. Impact of primary colorectal Cancer location on the KRAS status and its prognostic value. BMC Gastroenterol. 2019 Mar 27;19(1):46. doi: 10.1186/s12876-019-0965-5. PMID: 30917791; PMCID: PMC6437985.
- Watanabe T, Yoshino T, Uetake H, Yamazaki K, Ishiguro M, Kurokawa T, Saijo N, Ohashi Y, Sugihara K. KRAS mutational status in Japanese patients with colorectal cancer: results from a nationwide, multicenter, cross-sectional study. Jpn J Clin Oncol. 2013 Jul;43(7):706-12. doi: 10.1093/jjco/hyt062. Epub 2013 May 8. PMID: 23657052.
- Bollag G, Adler F, elMasry N, McCabe PC, Conner E Jr, Thompson P, McCormick F, Shannon K. Biochemical characterization of a novel KRAS insertion mutation from a human leukemia. J Biol Chem. 1996 Dec 20;271(51):32491-4. doi: 10.1074/jbc.271.51.32491. PMID: 8955068.
- Tong JH, Lung RW, Sin FM, Law PP, Kang W, Chan AW, Ma BB, Mak TW, Ng SS, To KF. Characterization of rare transforming KRAS mutations in sporadic colorectal cancer. Cancer Biol Ther. 2014 Jun 1;15(6):768-76. doi: 10.4161/cbt.28550. Epub 2014 Mar 18. PMID: 24642870; PMCID: PMC4049792.
- Huang, L., Guo, Z., Wang, F. et al. KRAS mutation: from undruggable to druggable in cancer. Sig Transduct Target Ther 6, 386 (2021). https://doi.org/10.1038/s41392-021-00780-4.
- Parikh, K., Banna, G., Liu, S.V. et al. Drugging KRAS: current perspectives and state-of-art review. J Hematol Oncol 15, 152 (2022).
- Janne PA, Riely GJ, Gadgeel SM, et al. Adagrasib in non-small-cell lung cancer harboring a KRAS(G12C) mutation. N Engl J Med. 2022;387(2):120–31.
Content Alerts
SignUp to our
Content alerts.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.