Biology Group. 2021 November 23;2(11):1121-1131. doi: 10.37871/jbres1357.
Virtual Screening of Phytochemicals Targeting the Main Protease and Spike Protein of SARS-CoV-2: An in silico Approach
Pallavi Gulati1, Aarti Yadav1, Jatin Chadha2 and Sandeepa Singh3*
2Department of Microbiology, Panjab University, Chandigarh, India
3Department of Botany, Maitreyi College, University of Delhi, New Delhi, India
- SARS-CoV-2
- COVID-19
- Main protease
- Spike protein
- Molecular docking
- Bioactive phytochemicals
Abstract
Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) is an emerging virus responsible for the ongoing Coronavirus Disease 19 (COVID-19) pandemic. Despite the advent of COVID-19 vaccines, pandemic fatigue is still escalating as new SARS-CoV-2 variants emerge and vaccine shortages hit globally. Hence, drug repurposing remains an alternative strategy to combat SARS-CoV-2. For centuries, plants have served as natural reservoirs of pharmacologically active compounds with minimal cytotoxicity and promising antimicrobial and antiviral activities. In this light, the present study was undertaken to virtually screen 33 phytochemicals across various cultivars against the main protease (Mpro) and Spike (S) protein of SARS-CoV-2 using ADME analysis. 31 phytochemicals obeying Lipinski’s rules were subjected to molecular docking using AutoDock Vina. Docking scores were determined by selecting the best conformation of the protein-ligand complex that exhibited the highest affinity. The study identified withanone, licoflavone A, and silibinin to interact with the S protein at the hACE2-binding site with high binding energies. Similarly, myricitrin, withanone, naringenin, licoflavone A, and silibinin exhibited high binding affinities with the substrate-binding pocket of Mpro between the domains I and II. Interestingly, licoflavone A, silibinin, and withanone interacted with both Mpro and S proteins in silico. Further, drug-likeness studies indicated withanone to be the most readily bioavailable phytochemicals among the three shortlisted ligands. Therefore, phytochemicals can be regarded as potential leads for developing inhibitors against this mysterious virus. In vitro investigations are further warranted to prove their antiviral efficacy.
Abbreviations
ADME: Absorption Distribution Metabolism Excretion; COVID-19: Coronavirus Disease-19; hACE2: Human Angiotensin-Converting Enzyme 2 Receptor; Mpro: Main Protease; NTD: N-terminal Domain (NTD); RBD: Receptor-Binding Domain (RBD); RBM: Receptor-Binding Motif; S protein: Spike Protein; SARS-CoV-2: Severe Acute Respiratory Syndrome Coronavirus-2; WHO: World Health Organization
Introduction
The past year has witnessed a severe collapse of the global healthcare system and downturned leading economies, disrupting livelihoods, impacting all trade sectors, every individual in every part of the world [1]. All these were repercussions following the emergence and widespread dissemination of the Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), resulting from the outbreak of Coronavirus Disease 19 (COVID-19) pandemic. This enigmatic pathogen has been regarded as a re-emerging virus that seriously risks human life [2]. SARS-CoV-2 is an enveloped virus displaying characteristic crown-like projections on its surface. Structurally, the virus codes for four structural proteins, namely the Envelope (E), Membrane (M), Spike (S), and Nucleocapsid (N) proteins [3]. These proteins cumulatively aid viral entry into host cells, subsequent infection, replication, and assembly of virion particles. The first outbreak of SARS-CoV-2 was reported in Wuhan, China, and since then, SARS-CoV-2 has spread across 220 countries, infecting more than 184 million people and resulting in over 3.9 million deaths [4]. Owing to its highly mutative positive-sense ssRNA genome, infection cycle, and the expansion of viral geographical range, variants of SARS-CoV-2 have also emerged during this pandemic [3]. This makes the present situation more intense, making the epidemiologists and researchers reiterate their line of action before the mutants unleash a wave of violent infections. Although six COVID-19 vaccines have been approved by the World Health Organization (WHO), their long-term efficacy against the upcoming SARS-CoV-2 variants remains a question. It is, therefore, the need of the hour to develop effective therapeutics against this deadly virus. Drug repurposing seems to be an alternative in this battleground as it bypasses the need for drug development, thereby reducing the drug discovery time frame. However, the existing repurposed drugs have been sidelined by the scientific community due to either lack of concrete evidence or associated side effects [5]. To overcome these shortcomings, naturopathic-based therapies have gained prominence among scientists. This involves using plant-derived formulations and herbal products that boast high target specificity, boosting the immune system with minimal cytotoxicity. As such, plants are known to harbor a rich repository of secondary metabolites such as alkaloids, flavonoids, terpenoids, tannins, saponins, and polyphenols with the ability to resist pathogens and boost host immunity [6]. These plant bioactives must therefore be investigated for their antiviral properties against the SARS-CoV-2. In recent times, bioinformatics has played an integral role in shaping scientific advancements by reducing the time and monetary costs imposed by experimental designs, execution, data analysis, lead selection, and laboratory-based trials. In this regard, molecular docking has proved to be an innovative algorithmic-based scientific tool for modeling and identifying characteristic interactions between the target protein and ligand/drug molecule in silico.
Several research groups have identified phytochemicals exhibiting antiviral properties against poliovirus, influenza virus, dengue virus, and even the Human Immunodeficiency Virus (HIV) by inhibiting either cell entry, attachment to a cell receptor, or viral replication [7]. Rhizome extracts of Curcuma longa (turmeric) have been shown to inhibit the activity of influenza A neuraminidases [8] and H5N1 avian influenza virus [9]. Carica papaya (papaya) demonstrates potential antiviral activity against Dengue virus with reduced thrombocytopenia and iMproved immunity during dengue fever [10]. In contrast, Zingiber officinale (ginger) has proved to be effective against the human respiratory syncytial virus [11] and Chikungunya virus [12]. in silico studies have recently identified potential antiviral drugs combating SARS-CoV-2 by targeting its structural proteins [13] or those involved in viral replication and assembly [14]. The crucial determinants of SARS-CoV-2 are its structural proteins, particularly the S protein that facilitates attachment to the host cells via the human Angiotensin-Converting Enzyme 2 (hACE2) receptor [15]. It lays the principle foundation for SARS-CoV-2 infection as it mediates the process of virion attachment and entry. The S protein contains two subunits, the S1 and S2; further distinguished into various functional domains. The S1 subunit includes the N-Terminal Domain (NTD) and the Receptor-Binding Domain (RBD), which houses the Receptor-Binding Motif (RBM). In the process of virus attachment, the S protein undergoes conformational changes resulting in endosome-mediated entry into the host cells [3]. Any amino acid substitutions (mutations) in the S protein can result in SARS-CoV-2 variants with an enhanced affinity towards the hACE2 receptor. Therefore, the S protein of SARS-CoV-2 becomes the primary target for abrogating viral attachment to human cells.
Another unique determinant of SARS-CoV-2 is its main protease (Mpro), or chymotrypsin-like protease (3CLpro) that controls the replication complex by catalyzing the cleavage of polyproteins translated from viral RNA, giving rise to functional viral proteins [16]. It also participates in the deubiquitination of cellular proteins and ISG15 removal, thereby causing immune suppression in the host cells [17]. Hence, Mpro seems to be an attractive antiviral target holding the key to replication machinery in SARS-CoV-2. Considering the quintessential role of S protein and Mpro in SARS-CoV-2 biology and virulence, this study was undertaken to screen phytochemicals against these target proteins. Using ADME analysis coupled with in silico docking analysis, we investigated the possible interactions between the S protein and Mpro of SARS-CoV-2 with 33 test phytochemicals based on their binding energies and docking scores. This adds to the volume of existing literature, thereby harnessing the antiviral potential of plant-derived secondary metabolites in combating SARS-CoV-2. Nevertheless, further bench-based investigations are warranted to substantiate these findings for their large-scale utility and viability.
Materials and Methods
Sources of ligands and viral target proteins
33 phytochemicals were selected for this study based on the previously published scientific literature on plant-derived antivirals [7,18]. The three-Dimensional (3D) structures of the ligands were retrieved from the PubChem database in SDF format. PubChem is a database that harbors the 3D structures and chemical information of organic compounds [19]. The downloaded SDF files were converted to PDB format using PyMOL. The phytochemicals used in the present study have been mentioned in table 1. Ribavirin, a guanosine analog that inhibits RNA/DNA replication [20], and ivermectin, an FDA-approved anti-parasitic drug and inhibitor of SARS-CoV-2 [21] were chosen as positive controls for this study against Mpro and S proteins, respectively. Similarly, two viral target proteins: Mpro and S protein were selected for docking studies. The Mpro and S protein crystal structures were retrieved in PDB format from RCSB PDB for the present study (www.rcsb.org). SARS-CoV-2 Mpro bound to N3 inhibitor (PDB ID: 6LU7) and RBD of S protein complexed with hACE2 (PDB ID: 6M0J) were downloaded for in silico analysis.
ADME analysis
Phytochemicals were initially screened to eliminate the compounds violating Lipinski’s rule of five [22]. The pharmacokinetic properties such as Absorption, Distribution, Metabolism, and Excretion (ADME) of the phytochemicals were determined using the SwissADME tool [23]. The phytochemicals in the PDB format were converted to standard SMILES format and uploaded to the SwissADME prediction tool (http://www.swissadme.ch). Pharmacological properties such as drug solubility (logS), Human Intestinal Absorption (HIA), and any carcinogenic traits were obtained from AdmetSAR (http://lmmd.ecust.edu.cn/admetsar2). The work plan for this study has been presented in figure 1.
Ligand and receptor preparation
The downloaded 3D structures were checked for the presence of any iMproper bonds, side-chain anomalies, and missing hydrogens using PyMOL software [24]. The water molecules, complex molecules, ions, and protein ligands were also removed from the 3D structures. For simplifying docking analysis, polar hydrogens were then added along with gagster charges prior to docking. The optimized PDB structures of Mpro, RBD spike, and all phytochemicals were finally uploaded and converted to PDBQT format via the AutoDock-MGT Tool.
Molecular docking, analysis, and visualization
Molecular Docking was carried out using AutoDock Vina 1.5.6, a graphical user interface used for predicting the receptor-ligand binding. AutoDock Vina is the successor of AutoDock 4 and is more robust, accurate, and user-friendly than its earlier version. Docking protocol was followed as described previously [25]. In the case of Mpro, targeted docking was performed with grid box dimensions 40 Å × 40 Å × 40 Å with grid spacing 0.7 Å, whereas in the case of RBD, spike molecule grid box dimensions were set to 52Å × 86 Å × 40 Å and grid spacing 0.6 Å. The value of exhaustiveness was set at 24. Each docking experiment resulted in nine confirmations with their respective binding affinity in kcal/mol. The ligand-binding pose with the lowest binding energy was recruited for further studies. To confirm the fidelity of docking software, ligands were re-docked to the Mpro and S protein crystal structures. The protein-ligand interactions and PDB structures were viewed and analyzed in PyMOL molecular visualization tool [24] and LigPlot [26]. The phytochemicals found to interact with both the viral target proteins intensely were further discussed in our study.
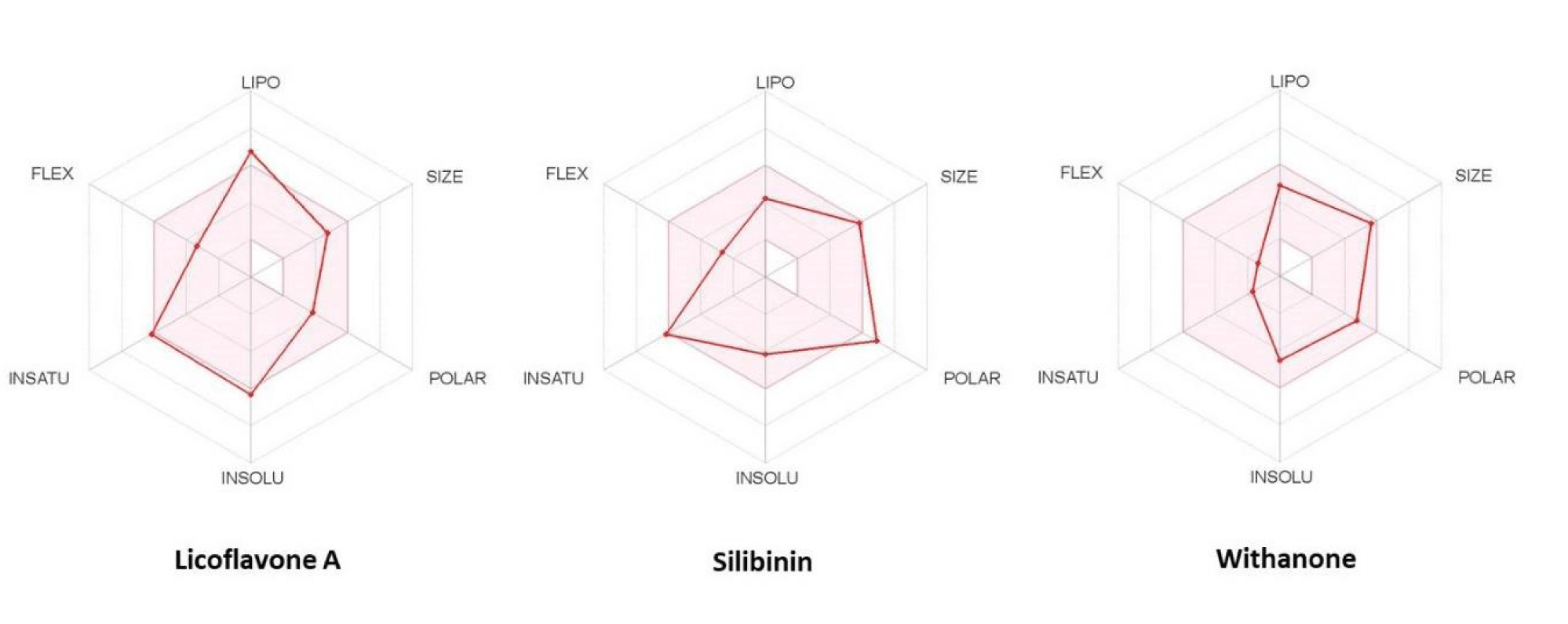
Bioavailability radar
The drug-likeness of the shortlisted phytochemicals with binding energy lower than the controls were analyzed by considering six physiochemical properties and forming a bioavailability radar using the SwissADME tool (https://www.swissadme.ch/). Six parameters were evaluated: ligand flexibility, lipophilicity, polarity, saturation, size, and solubility. The pink-shaded regions define the optimal values of the six parameters, and any deviation from them suggests the ligand not being orally bioavailable [23].
Results and Discussion
The unprecedented outbreak of COVID-19 has created a global threat and prompted scientists worldwide to develop efficacious antiviral therapies with high target specificity and minimal off-target effects. There is a pressing need to replace antiviral-based therapies due to many factors, including the emergence of resistance due to high mutation rates, virus latency, non-specificity or coMpromised target specificity, undesired side effects, poor outcomes in clinical setups. Moreover, poor drug compliance and untimely administration of antivirals are other significant reasons for the failure of antiviral drugs. To the rescue, plant extracts harboring numerous bioactive phytochemicals have proved effective in combating human viral diseases [27]. Numerous studies have shown that these nature-derived products exhibit robust antiviral activity since they bind to critical viral proteins, impeding virus replication, even at nanomolar concentrations [28]. Most of these phytochemicals are produced by plants as secondary metabolites to fend off microorganisms and thus can be repurposed as potential drug candidates against SARS-CoV-2. The roots of plant-based elixirs can be traced to Ayurveda, Siddha, and Unani, the ancient healthcare systems of India [6]. Their usage has been faithfully transmitted for ages between different generations. However, general disagreements and ambiguity related to their use in the current scenario pertaining to the lack of knowledge regarding their mode of action persist. In this view, our study was aimed to screen phytochemicals and their possible role in targeting two essential proteins of SARS-CoV-2 using computational analysis. It is well known that the main protease Mpro is vital for polyprotein processing, and spike plays a crucial role in mediating viral attachment to host cells [29]. Because of their essential role in regulating the viral life cycle and absence of any human orthologs counterpart, the two viral proteins were considered attractive drug targets against the SARS-CoV-2.
ADME analysis identified 31 phytochemicals to obey Lipinski’s rules
Lipinski’s rule of five was applied to all selected 33 candidates to shortlist ligands based on their physicochemical properties and drug-likeness. These rules enlist various parameters for a chemical that should be satisfied before its declaration as an oral drug. It helps determine the compound's suitability as an active drug for oral consumption in humans [22]. These parameters require molecular weight to be less than 500 Da, the number of hydrogen bond acceptors and donors to be less than 10 and 5, respectively, and lipophilicity (log P), i.e., an octanol-water coefficient to be less than 5 [22]. It has been well documented that molecules that fail to follow Lipinski’s rule have poor absorption and bioavailability in the body. Therefore, compounds that violated more than two parameters were ruled out, and other compounds were considered for further studies and analysis. Out of the 33 phytochemicals evaluated; two compounds: procyanidin B2 and liquiritin apioside, violated more than two parameters with very low lipophilicity and high numbers of H-bond donors and acceptors (Table 1). Henceforth, the remaining 31 compounds were subjected to molecular docking.
Phytochemicals exhibit a high binding affinity towards viral proteins
With the aim to inhibit viral replication and impede its attachment to host cells, we screened a series of phytochemicals to target the Mpro and S proteins, respectively. Molecules that could bind effectively to the substrate-binding pocket of Mpro or could sandwich between hACE2 receptor and S protein were speculated to be therapeutic leads. Recent computational investigations have scrutinized various phytochemicals that effectively bind Mpro and S proteins with high affinity [24,30]. To extend previous findings, the 31 shortlisted phytochemicals were examined for their abilities to interact with the target proteins in silico. We employed AutoDock Vina to analyze the binding affinities, position, and conformation of ligands concerning the viral target proteins. Binding energy, measured in kcal/mol, is the sum of intermolecular interactions between the test protein and ligands. It is inversely proportional to affinity, i.e., the lower the binding energy of a test ligand, the higher is its affinity for the protein [31]. The docking results showed multiple conformations of ligand (pose) with binding energies ranging from -9 kcal/mol (myricitrin) to -3.4 kcal/mol (betaine). It has been generally recommended that a binding affinity lesser than the upper threshold of − 6 kcal/mol be regarded as the cut-off value for deciding a strong interaction between the ligand and target protein [24]. Among the 31 docked ligands, 18 compounds displayed a higher binding affinity towards Mpro than ribavirin (-6.1 kcal/mol). Five compounds showed increased binding affinity more than ivermectin (-7.3 kcal/mol) for the S protein, while 15 compounds exhibited values lower than -6 kcal/mol. The binding energies of the test phytochemicals for Mpro and S protein have been depicted in figure 2.
Molecular docking and interactions of phytochemicals with SARS-CoV-2 Mpro
It has been well documented that Mpro is an essential protease of SARS-CoV-2 involved in virus replication, aiding post-translational polyprotein processing [32]. Thus, it has been utilized as an attractive drug target in numerous studies. The 3D structure of Mpro accommodates three regions: domain I and domain II consisting of a series of beta barrels, whereas domain III is composed of alpha-helices. The active site of Mpro is positioned between domain I and domain II flanked by the His41-Cys145 catalytic dyad [33,34]. Catalytic dyad is found in various enzymes that utilize cysteine as the nucleophile deprotonated by the -NH group of histidine, leading to oxyanion hole formation. The latter is eventually stabilized by hydrogen bond donors: Gly143, Ser144, and Cys145 of Mpro [33]. Moreover, mutations in the catalytic dyad residues have been to abolish the activity of Mpro [35]. Thus, targeted docking was carried out wherein the docking region was centered on the respective coordinates at the interface between the two domains such that it encompasses the whole substrate-binding pocket of Mpro. Virtual docking revealed that most phytochemicals occupied similar positions within the active site, located between the domains I and II (Figure 3a). Amino acids Arg188, Gln189, Met49, Cys145, His41, His163, His164, and Met165 were predicted to be the critical residues of Mpro majorly involved in interactions with the test phytochemicals (Figure 3b).
Notably, myricitrin was best fitted into the active site pocket of Mpro by two hydrogen bonds with Tyr54 and Asp187 via the phenyl ring of the flavonoid backbone (Figure 3b). The sugar residues also formed four hydrogen bonds with Leu141, Asn142, Ser144, and His163. Apart from this, it also interacted with Mpro through a plethora of hydrophobic bonds, including His41, Gly143, Cys145, His164, Met165, Glu166, Arg188, and Gln189. Similarly, licoflavone A was enveloped by 12 hydrophobic bonds, including Thr25, Thr26, His41, Met49, Leu141, Asn142, His163 Met165, Pro168, Arg188, Gln189, and Thr190 (Figure 3b). Moreover, the phenyl of ring of the flavonoid backbone formed one hydrogen bond with Cys145 and Gly143 and two hydrogen bonds by Ser144. Also, both the phenyl ring and heterocyclic ring of naringenin were hydrogen-bonded with Leu141, Ser144, Gly143, Glu166, and Asp187. Likewise, numerous other phytochemicals were surrounded by a hydrophobic environment and hydrogen bonds that could stabilize their interactions with the binding pocket of Mpro. Briefly, naringenin, withanone, silibinin, ursolic acid, oleanolic acid, rosmarinic acid, stigmasterol, and E-guggulsterone were reportedly stabilized by 8, 11, 9, 9, 9, 7, 8, and 4 hydrophobic bonds, respectively. In contrast, the latter phytochemicals showed 5, 3, 4, 3, 1, 7, 2, and 2 hydrogen bonds, respectively.
Myricitrin showed maximum affinity with the remarkable binding energy of -9 kcal/mol among all the test phytochemicals. Myricitrin is a glycosyloxyflavone well known for its anti-inflammatory and antioxidant properties [36]. Furthermore, Gao et al. reported the anti-atherosclerotic and hypolipidemic effects of myrcitirin [37]. Pentacyclic triterpenoids such as α-boswellic acid, oleanolic acid, and ursolic acid also showed excellent binding energies (lower than -6 kcal/mol) with numerous interactions. The antiviral, antitumor and antimicrobial properties of triterpenoids have also been documented [38]. Oleanolic acid, present in olive oil, is also known for its immunomodulatory effects and is widely recognized in cancer prevention [39]. Also, E-guggulsterone derived from gum resin of Commiphora wightii, demonstrated superior binding due to numerous interactions with Mpro compared to ribavirin.
Interestingly, most phytochemicals that showed high affinity or low binding energies and intermolecular interactions towards Mpro were flavonoids such as myricitin, naringenin, and licoflavone A. Their binding sites were consistent with ketoamide, a broad-spectrum inhibitor of Coronavirus, flanked by the His-Cys catalytic dyad [40]. Therefore, five lead compounds that showed a high binding affinity for Mpro with solid intermolecular interactions were myricitrin, withanone, licoflavone A, silibinin, and naringenin. Interestingly the majority of these compounds are flavonoids. Thus, it can be speculated that compounds containing flavonoids can serve as potent inhibitors of Mpro.
Molecular docking reveals high-affinity interaction between phytochemicals and S protein
The prime phase in infection by SARS-CoV-2 is mediated by its S protein, aiding in recognizing the hACE2 receptor and subsequent fusion to the host cell membrane [15]. Moreover, the upcoming wave of infections has been closely related to the selection of SARS-CoV-2 variants due to amino acid substitutions in the S protein. Hence, targeting the S protein seems to be the most promising remedy for combating SARS-CoV-2 in the long run. The S protein of SARS-CoV-2 is a transmembrane homo-trimeric protein with two distinct domains. The S1 domain bears the RBD and binds to the host cell receptor, while the highly conserved S2 domain regulates viral fusion with the host cell membrane [41]. Thus, docking was targeted to strictly restrict phytochemicals around the hACE2 receptor-directing face of the RBD, such that the interactions between S protein and hACE2 receptor are disrupted. Virtual screening revealed that most ligands (18) exhibit binding affinities lower than -6 kcal/mol, with five phytochemicals demonstrating binding energies superior to ivermectin (-7.3 kcal/mol) and multiple interactions with the RBD (Figure 2). These include α-boswellic acid, licoflavone A, oleanolic acid, ursolic acid, and withanone.
Similar to Mpro, interactions were predominantly stabilized by hydrophobic bonds with the RBD spike, followed by hydrogen bonds. Interestingly, hydrophobic bonds outnumbered the hydrophobic bonds in all cases, except with rosmarinic acid. Some of the critical residues on S protein that played a major role in interactions were Tyr505, Tyr495, and Arg408. Withanone displayed the highest affinity (-8.4 kcal/mol) with the nine interactions, while silibinin displayed the maximum interactions (eleven) on the S protein and hACE2 receptor interface. Briefly, withanone formed hydrophobic bonds with Thr415, Gly416, Lys417, Tyr453, Leu465, and hydrogen bonds with Glu406 and Gln409 Thr415, and Arg401 (Figures 4a). Due to its binding position, withanone was found wedged between the hACE2 receptor and S protein. Hence, it can be considered as a probable candidate for disrupting the virus-host cell interactions. Moreover, our findings are in agreement with the recent studies that regard withanone as a potential inhibitor of SARS-CoV-2 entry into host cells [42]. Withanone has been valued as an ancient medicinal herb in antistress therapy, memory enhancement, and nerve tonic with neuroprotective properties [43]. Its anticancerous properties have also been documented in the literature [44,45]. Hence, its potential application as an anti-COVID drug may be exploited to the core only after translating such in silico and bench-based research into clinical trials.
Although α-boswellic acid, oleanolic acid, and ursolic acid exhibited high affinities for the RBD, they were not taken into further consideration due to reduced binding to Mpro. Hence, we considered licoflavone A and silibinin as they exhibited excellent binding energies with numerous hydrophobic and hydrogen bonds (Figure 4b). Licoflavone A is known to cure gastric ulcers [46], while silibinin, a flavolingin, has renowned antineoplastic activity, alleviates inflammation, and induces apoptosis [47]. Licoflavone A was well fitted into the hACE2-binding face via hydrophobic interactions with Arg403, Phe497, Tyr495, Tyr453, and Leu455 residues along with hydrogen bonding with Tyr505, Asn501, and Gly 496. Similarly, silibinin was also well accommodated in the RBD by forming multiple hydrophobic bonds with Asn501, Gln498, Gly496, Arg403, Tyr495, Tyr453, Leu455, and Lys417 and hydrogen bonds with Tyr449, Ser494, and Glu406. Multiple hydrogen bonds between phytochemicals and target protein can be regarded to enhance the stability of interactions. Numerous studies have elaborated on the role of hydrophobic interactions in protein-ligand interaction. Patil, et al. [48] demonstrated that hydrophobic interactions and hydrogen bonds at the drug target interface play a vital role in stabilizing the intermolecular interactions and significantly impact drug efficacy. In the same direction, Freitas, et al. [49] highlighted the role of hydrophobic and hydrogen bonds in stabilizing the protein-ligand interactions in the proper orientation. Considering the high binding affinities and the myriad of hydrogen and hydrophobic bonds between the phytochemicals and S protein, it may help circumvent the menace of COVID-19. However, their ability to abrogate the hACE2-spike complex association needs further evaluation by in vitro investigations.
Bioavailability radar reveals withanone to be the most suitable phytochemical against Mpro and S protein
The drug-likeness of three shortlisted phytochemicals viz. silibinin, licoflavone A, and withanone was predicted using physicochemical and ADME properties determined from SwissADME [23]. SwissADME is an authenticated web tool for predicting and evaluating the pharmacokinetics, drug-likeness, and medicinal chemistry friendliness of small molecules. Its logarithm is built on Lipinski's rule of five [50] while presenting a user-friendly interface for non-experts in computer-aided drug design [23]. To be estimated as drug-like, the red line of the compound under study must be fully included in the pink area. Based on the results, it was observed that all three phytochemicals obeyed the molecular size, bond flexibility, and saturation parameters. We found only withanone to be orally bioavailable, fulfilling all the necessary parameters by entirely fitting in the pink-shaded area (Figure 5). On the other hand, licoflavone A and silibinin showed slight deviations from the lipophilicity and polarity (topological polar surface area) parameters and thus represented a suboptimal physicochemical property for oral bioavailability (Figure 5). Nevertheless, our results add to the expanding literature in this avenue for repurposing phytochemicals as anti-SARS-CoV-2 molecules (Figures S1 & S2).
Conclusion
The last two decades have witnessed the outbreak of emerging diseases, of which SARS-CoV-2 presently plagues humankind. Due to the rapid rate of mutations in its RNA genome, it has become even more challenging to design potential therapeutic drugs against them. Hence, repurposing traditional bioactive phytochemicals seems to be an immediate alternative at our disposal [51]. In the present study, 33 phytochemicals were selected across various cultivars and virtually screened for targeting the Mpro and S proteins of SARS-CoV-2. These compounds were screened using Lipinski’s rule of five and drug-likeness parameters. 31 compounds that obeyed Lipinski’s rules were subjected to molecular docking alongside ribavirin and ivermectin as controls. Several phytochemicals showed a high binding affinity for Mpro and S proteins individually, while only licoflavone A, silibinin, and withanone demonstrated high affinity for both the target proteins. Of the three phytochemicals, only withanone was found to be orally bioavailable, while licoflavone A and silibinin were sub-optimally bioavailable. Considering that plant extracts harbor several phytochemicals, they can be exploited to target this notorious virus in a multi-targeted approach that might have synergistic effects. Thus, the results obtained in the current study yield promising results in harnessing the ancient Indian knowledge of Ayurveda in treating human ailments, including SARS-CoV-2. Moreover, the rampant use of antibiotics in this COVID-19 era has led to a sudden “antibiotic rush” that can accelerate the pace at which resistance in bacterial pathogens evolves, severely coMpromises the global commitment to curb antimicrobial resistance and the incidence of secondary prevention infections in COVID-19 patients [52]. Thus, bioactive phytochemicals can prove to be the “wonder drugs” with multifaceted properties in this aspect as well. Until then, concrete evidence from in vitro and in vivo investigations is warranted to predict and validate the use of phytochemicals in clinical setups to defeat the ongoing COVID-19 pandemic.
Acknowledgment
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors. Financial support from the Indian Council of Medical Research (ICMR) for providing fellowship to JC is appreciated.
References
- Kaye AD, Okeagu CN, Pham AD, Silva RA, Hurley JJ, Arron BL, Sarfraz N, Lee HN, Ghali GE, Gamble JW, Liu H, Urman RD, Cornett EM. Economic impact of COVID-19 pandemic on healthcare facilities and systems: International perspectives. Best Pract Res Clin Anaesthesiol. 2021 Oct;35(3):293-306. doi: 10.1016/j.bpa.2020.11.009. Epub 2020 Nov 17. PMID: 34511220; PMCID: PMC7670225.
- Celik İ, SaatCİ E, & Eyuboglu FÖ. Emerging and reemerging respiratory viral infections up to Covid-19. Turk J Med Sci. 2020;50(SI-1):557-562. doi: 10.3906/sag-2004-126. PMID: 32293833.
- Chadha J, Khullar L, Mittal N. Facing the wrath of enigmatic mutations: a review on the emergence of severe acute respiratory syndrome coronavirus 2 variants amid coronavirus disease-19 pandemic. Environ Microbiol. 2021; Online ahead of print. doi: 10.1111/1462-2920.15687. PMID: 34320263.
- Worldometer COVID-19 Coronavirus Pandemic [Internet]. 2021.
- Galindez G, Matschinske J, Rose TD, Sadegh S, Salgado-Albarrán M, Späth J, Baumbach J and Pauling JK. Lessons from the COVID-19 pandemic for advancing computational drug repurposing strategies. Nat Comput Sci. 2021;1(1):33-41. doi: 10.1038/s43588-020-00007-6
- Chadha J, Gupta M, Nagpal N, Sharma M, Adarsh T, Joshi V, Tiku V, Mittal T, Nain VK, Singh A, Snigdha SK, Chandra NS, John S and Diwan P. Antibacterial potential of indigenous plant extracts against multidrug-resistant bacterial strains isolated from New Delhi region. GSC Biol Pharm Sci. 2021;14(2):185-196. doi: 10.30574/gscbps.2021.14.2.0053
- Naithani R, Huma LC, Holland LE, Shukla D, McCormick DL, Mehta RG, Moriarty RM. Antiviral Activity of Phytochemicals: A Comprehensive Review. Mini Rev Med Chem. 2008;8(11):1106-1133. doi: 10.2174/138955708785909943. PMID: 18855727.
- Dao TT, Nguyen PH, Won HK, Kim EH, Park J, Won BY, Keun-Oh W. Curcuminoids from Curcuma longa and their inhibitory activities on influenza A neuraminidases. Food Chem. 2012; 134(1):21-28. doi: 10.1016/j.foodchem.2012.02.015
- Sornpet B, Potha T, Tragoolpua Y, & Pringproa K. Antiviral activity of five Asian medicinal pant crude extracts against highly pathogenic H5N1 avian influenza virus. Asian Pac J Trop Med. 2017; 10(9):871-876. doi: 10.1016/j.apjtm.2017.08.010. PMID: 29080615
- Sarker MMR, Khan F, & Mohamed IN. Dengue Fever: Therapeutic Potential of Carica papaya L. Leaves. Front Pharmacol. 2021;12. doi: 10.3389/fphar.2021.610912. PMID: 33981215.
- Chang JS, Wang KC, Yeh CF, Shieh DE, & Chiang LC. Fresh ginger (Zingiber officinale) has anti-viral activity against human respiratory syncytial virus in human respiratory tract cell lines. J Ethnopharmacol. 2021;145(1):146-151. doi: 10.1016/j.jep.2012.10.043. PMID: 23123794.
- Kaushik S, Jangra G, Kundu V, Yadav JP, & Kaushik S. Anti-viral activity of Zingiber officinale (Ginger) ingredients against the Chikungunya virus. VirusDisease. 2020; 31(3):270-276. doi: 10.1007/s13337-020-00584-0. PMID: 32420412
- Dutta K, Elmezayen AD, Al-Obaidi A, Zhu W, Morozova OV, Shityakov S, Khalifa I. Seq12, Seq12m, and Seq13m, peptide analogues of the spike glycoprotein shows antiviral properties against SARS-CoV-2: An in silico study through molecular docking, molecular dynamics simulation, and MM-PB/GBSA calculations. J Mol Struct. 2021; 1246:131113. doi: 10.1016/j.molstruc.2021.131113. PMID: 34305174.
- Dutta K, Shityakov S, Morozova O, Khalifa I, Zhang J, Panda A and Ghosh C. Beclabuvir can inhibit the RNA-dependent RNA polymerase of newly emerged novel Coronavirus (SARS-CoV-2). Preprints 2020; 2020030395. doi: 10.20944/preprints202003.0395.v1
- Huang Y, Yang C, Xu X-f, Xu W, Liu SW. Structural and functional properties of SARS-CoV-2 spike protein: Potential antivirus drug development for COVID-19. Acta Pharmacol Sin. 2020; 41(9):1141-1149. doi: 10.1038/s41401-020-0485-4
- Jin Z, Du X, Xu Y, Deng Y, Liu M, Zhao Y, Zhang B, Li X, Zhang L, Peng C, Duan Y, Yu J, Wang L, Yang K, Liu F, Jiang R, Yang X, You T, Liu X, Yang X, Bai F, Liu H, Liu X, Guddat LW, Xu W, Xiao G, Qin C, Shi Z, Jiang H, Rao Z, Yang H. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020; 582(7811):289-293. doi: 10.1038/s41586-020-2223-y. PMID: 32272481
- Shin D, Mukherjee R, Grewe D, Bojkova D, Baek K, Bhattacharya A, Schulz L, Widera M, Mehdipour AR, Tascher G, Geurink PP, Wilhelm A, van der Heden van Noort GJ, Ovaa H, Müller S, Knobeloch KP, Rajalingam K, Schulman BA, Cinatl J, Hummer G, Ciesek S, Dikic I. Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity. Nature. 2020; 587(7835):657-662. doi: 10.1038/s41586-020-2601-5. PMID: 32726803.
- Shabat S, Yarmolinsky L, Porat D, & Dahan A. Antiviral effect of phytochemicals from medicinal plants: Applications and drug delivery strategies. Drug Deliv Transl Res. 2019; 10(2):354-367. doi: 10.1007/s13346-019-00691-6. PMID: 31788762.
- Wang Y, Xiao J, Suzek TO, Zhang J, Wang J, Bryant SH. PubChem: a public information system for analyzing bioactivities of small molecules. Nucleic Acids Res. 2009; 37:623-633. doi: 10.1093/nar/gkp456. PMID: 19498078.
- Celik I, Erol M, & Duzgun Z. in silico evaluation of potential inhibitory activity of remdesivir, favipiravir, ribavirin and galidesivir active forms on SARS-CoV-2 RNA polymerase. Mol Divers. 2021. doi: 10.1007/s11030-021-10215-5. PMID: 33765239.
- Caly L, Druce JD, Catton MG, Jans DA, & Wagstaff KM. The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antiviral Res. 2020;178:104787. doi: 10.1016/j.antiviral.2020.104787. PMID: 32251768.
- Lipinski CA. Lead- and drug-like compounds: the rule-of-five revolution. Drug Discov Today Technol. 2004; 1(4):337-341. doi: 10.1016/j.ddtec.2004.11.007. PMID: 24981612.
- Daina A, Michielin O, Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep. 2017; 7(1). doi: 10.1038/srep42717
- Hiremath S, Kumar HDV, Nandan M, Mantesh M, Shankarappa KS, Venkataravanappa V, Basha CRJ, Reddy CNL. in silico docking analysis revealed the potential of phytochemicals present in Phyllanthus amarus and Andrographis paniculata, used in Ayurveda medicine in inhibiting SARS-CoV-2. 3 Biotech. 2021; 11(2). doi: 10.1007/s13205-020-02578-7. PMID: 33457171.
- Forli S, Huey R, Pique ME, Sanner MF, Goodsell DS, Olson AJ. Computational protein–ligand docking and virtual drug screening with the AutoDock suite. Nat Protoc. 2016; 11(5):905-919. doi: 10.1038/nprot.2016.051. PMID: 27077332.
- Wallace AC, Laskowski RA, & Thornton JM. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 1995; 8(2):127-134. doi: 10.1093/protein/8.2.127. PMID: 7630882.
- Mukhtar M, Arshad M, Ahmad M, Pomerantz RJ, Wigdahl B, Parveen Z. Antiviral potentials of medicinal plants. Virus Res. 2008; 131(2):111-120. doi: 10.1016/j.virusres.2007.09.008. PMID: 17981353.
- Islam MT, Sarkar C, El-Kersh DM, Jamaddar S, Uddin SJ, Shilpi JA, Mubarak MS. Natural products and their derivatives against coronavirus: A review of the non‐clinical and pre‐clinical data. Phytother Res. 2020; 34(10):2471-2492. doi: 10.1002/ptr.6700. PMID: 32248575.
- Gioia M, Ciaccio C, Calligari P, De Simone G, Sbardella D, Tundo G, Fasciglione GF, Di Masi A, Di Pierro D, Bocedi A, Ascenzi P, Coletta M. Role of proteolytic enzymes in the COVID-19 infection and promising therapeutic approaches. Biochem Pharmacol. 2020; 182:114225. doi: 10.1016/j.bcp.2020.114225. PMID: 32956643.
- Garg S, Anand A, Lamba Y, & Roy A. Molecular docking analysis of selected phytochemicals against SARS-CoV-2 Mpro receptor. Vegetos. 2020; 33(4):766-781. doi: 10.1007/s42535-020-00162-1. PMID: 33100613.
- Pantsar T & Poso A. Binding Affinity via Docking: Fact and Fiction. Molecules. 2018; 23(8):1899. doi: 10.3390/molecules23081899. PMID: 30061498.
- Xue X, Yu H, Yang H, Xue F, Wu Z, Shen W, Li J, Zhou Z, Ding Y, Zhao Q, Zhang XC, Liao M, Bartlam M, Rao Z. Structures of Two Coronavirus Main Proteases: Implications for Substrate Binding and Antiviral Drug Design. J Virol. 2008; 82(5):2515-2527. doi: 10.1128/JVI.02114-07. PMID: 18094151.
- Chou CY, Chang HC, Hsu WC, Lin TZ, Lin CH, Chang GG. Quaternary structure of the severe acute respiratory syndrome (SARS) coronavirus main protease. Biochemistry. 2010; 115-128. doi: 10.1007/978-3-642-03683-5_8. PMID: 15554703.
- Kneller DW, Phillips G, O'Neill HM, Jedrzejczak R, Stols L, Langan P, Joachimiak A, Coates L, Kovalevsky A. Structural plasticity of SARS-CoV-2 3CL Mpro active site cavity revealed by room temperature X-ray crystallography. Nat Commun. 2020; 11(1). doi: 10.1038/s41467-020-16954-7. PMID: 32581217.
- Richter F, Blomberg R, Khare SD, Kiss G, Kuzin AP, Smith AJ, Gallaher J, Pianowski Z, Helgeson RC, Grjasnow A, Xiao R, Seetharaman J, Su M, Vorobiev S, Lew S, Forouhar F, Kornhaber GJ, Hunt JF, Montelione GT, Tong L, Houk KN, Hilvert D, Baker D. Computational Design of Catalytic Dyads and Oxyanion Holes for Ester Hydrolysis. J Am Chem Soc. 2012; 134(39):16197-16206. doi: 10.1021/ja3037367. PMID: 22871159.
- Domitrović R, Rashed K, Cvijanović O, Vladimir-Knežević S, Škoda M, Višnić A. Myricitrin exhibits antioxidant, anti-inflammatory and antifibrotic activity in carbon tetrachloride-intoxicated mice. Chem Biol Interact. 2015; 230:21-29. doi: 10.1016/j.cbi.2015.01.030. PMID: 25656916.
- Gao Y, Yan L, Huang Y, Liu F, Zhao Y, Cao L, Wang T, Sun Q, Ming Z, Zhang L, Ge J, Zheng L, Zhang Y, Wang H, Zhu Y, Zhu C, Hu T, Hua T, Zhang B, Yang X, Li J, Yang H, Liu Z, Xu W, Guddat LW, Wang Q, Lou Z, Rao Z. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020; 368(6492):779-782. doi: 10.1126/science.abb7498. PMID: 32277040.
- Xiao S, Tian Z, Wang Y, Si L, Zhang L, Zhou D. Recent progress in the antiviral activity and mechanism study of pentacyclic triterpenoids and their derivatives. Med Res Rev. 2018; 38(3):951-976. doi: 10.1002/med.21484. PMID: 29350407.
- Ríos JL. Effects of triterpenes on the immune system. J Ethnopharmacol. 2010; 128(1):1-14. doi: 10.1016/j.jep.2009.12.045. PMID: 20079412.
- Zhang L, Lin D, Sun X, Curth U, Drosten C, Sauerhering L, Becker S, Rox K, Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of iMproved α-ketoamide inhibitors. Science. 2020; 368(6489):409-412. doi: 10.1126/science.abb3405. PMID: 32198291.
- Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020; 181(2):281-292. doi: 10.1016/j.cell.2020.02.058. PMID: 32155444.
- Balkrishna A, Pokhrel S, Singh H, Joshi M, Mulay VP, Haldar S, Varshney A. Withanone from Withania somnifera Attenuates SARS-CoV-2 RBD and Host ACE2 Interactions to Rescue Spike Protein Induced Pathologies in Humanized Zebrafish Model. Drug Des Devel Ther. 2021; 15:1111-1133. doi: 10.2147/DDDT.S292805. PMID: 33737804.
- Dar NJ, Ahmad M. Neurodegenerative diseases and Withania somnifera (L.): An update. J Ethnopharmacol. 2020; 256:112769. doi: 10.1016/j.jep.2020.112769
- Gao R, et al. Withanone-Rich Combination of Ashwagandha Withanolides Restricts Metastasis and Angiogenesis through hnRNP-K. Mol Cancer Ther. 2014; 13(12):2930-2940. doi: 10.1158/1535-7163.MCT-14-0324. PMID: 32240781.
- Wadegaonkar VP & Wadegaonkar PA. Withanone as an inhibitor of survivin: A potential drug candidate for cancer therapy. J Biotechnol. 2013;168(2):229-233. doi: 10.1016/j.jbiotec.2013.08.028. PMID: 23994265.
- Yang Y, Wang S, Bao YR, Li TJ, Yang GL, Chang X, Meng XS. Anti-ulcer effect and potential mechanism of licoflavone by regulating inflammation mediators and amino acid metabolism. J Ethnopharmacol. 2017;199:175-182. doi: 10.1016/j.jep.2017.01.053. PMID: 28159726.
- Tong WW, Zhang C, Hong T, Liu DH, Wang C, Li J, He XK, Xu WD. Silibinin alleviates inflammation and induces apoptosis in human rheumatoid arthritis fibroblast-like synoviocytes and has a therapeutic effect on arthritis in rats. Sci Rep. 2018; 8(1). doi: 10.1038/s41598-018-21674-6. PMID: 29459717.
- Patil R, Das S, Stanley A, Yadav L, Sudhakar A, Varma AK. Optimized hydrophobic interactions and hydrogen bonding at the target-ligand interface leads the pathways of drug-designing. PLoS One. 2010;5(8):e12029. doi: 10.1371/journal.pone.0012029. PMID: 20808434.
- Freitas R & Schapira M. A systematic analysis of atomic protein–ligand interactions in the PDB. MedChemComm. 2017; 8(10):1970-1981. doi: 10.1039/c7md00381a
- Lipinski CA, Lombardo F, Dominy BW, & Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev. 2001;46(1-3):3-26. doi: 10.1016/s0169-409x(00)00129-0. PMID: 11259830.
- Chadha J, Harjai K and Chhibber S. Revisiting the virulence hallmarks of Pseudomonas aeruginosa: a chronicle through the perspective of quorum sensing. Environ Microbiol. 2021; Online ahead of print. doi: 10.1111/1462-2920.15784. PMID: 34559444.
- Chadha J, Harjai K and Chhibber S. Repurposing phytochemicals as anti-virulent agents to attenuate quorum sensing-regulated virulence factors and biofilm formation in Pseudomonas aeruginosa. Microb Biotechnol. 2021; Online ahead of publication. doi: 10.1111/1751-7915.13981
Content Alerts
SignUp to our
Content alerts.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.