Medicine Group . 2022 May 25;3(5):602-606. doi: 10.37871/jbres1487.
Pan Cancer Insights into BAZ1B as a Prognostic and Immunological Marker
Xiaorong Zhao1, Yuhu Niu1, Qingjuan Li1, Yuxuan Zhang1 and Bo Niu1,2*
2Department of Biotechnology, Beijing Municipal Key Laboratory of Child Development and Nutriomics, Capital Institute of Pediatrics, Beijing, China
- BAZ1B
- Pan-cancer
- Prognosis
- Immune
Abstract
Background: The Bromodomain Adjacent to the Zinc Finger Domain 1B (BAZ1B) is a versatile nuclear protein. BAZ1B acts on chromatin remodeling, participates in DNA replication and damage repair, and is involved in RNA transcription based on its kinase activity. Although many experimental data support a close relationship between BAZ1B and cancer, its role in pan-cancer remains unclear. Our study aimed to analyze the prognostic and immune functions of BAZ1B expression in different cancers.
Methods: Based on different databases, we investigated the potential carcinogenicity of BAZ1B, including the expression of all TCGA tumors, correlation with prognosis, Tumor Microenvironment (TME), protein phosphorylation, and functional enrichment.
Results: The results showed that BAZ1B was overexpressed in a variety of cancers, and there was a significant correlation between gene mutations and poor prognosis associated with high expression in most cancer patients. We observed elevated phosphorylation levels in breast cancer and lung adenocarcinoma. A significant relationship was also found between BAZ1B expression and TME, including tumor-associated fibroblast infiltration. We also observed that the cellular function of DNA and hepatocellular carcinoma may be related to the molecular mechanism of BAZ1B.
Conclusion: The prognostic and immunological markers of BAZ1B provides a new understanding for future studies in our first pan-cancer analysis.
Abbreviations
ACC: Adrenocortical Carcinoma; BAZ1B: Bromodomain Adjacent To Zinc Finger Domain 1B; BLCA: Bladder Urothelial Carcinoma; BP: Biological Process; BRCA: Breast Invasive Carcinoma; CC: Cellular Component; CESC: Cervical Squamous Cell Carcinoma and Endocervical Adenocarcinoma; CHOL: Cholangiocarcinoma; CI: Confidence Intervals; COAD: Colon Adenocarcinoma; DAVID: Database For Annotation, Visualization, and Integrated Discovery; DFS: Disease-Free Survival; DLBC: Lymphoid Neoplasm Diffuse Large B-Cell Lymphoma; DSS: Disease-Specific Survival; ESCA: Esophageal Carcinoma; FC: Fold Change; GBM: Glioblastoma Multiforme; GEO: Gene Expression Omnibus; GEPIA2: Interactive Analysis of Gene Expression Profile, 2nd Edition; GO: Gene Ontology; HCC: Hepatocllular Carcinoma; HNSC: Head And Neck Squamous Cell Carcinoma; HR: Hazard Ratio; ICPI: Immune Checkpoint Inhibitor; IL-2: Interleukin 2; KEGG: Kyoto Genome and Genome Encyclopedia; KICH: Kidney Chromophobe; KIRC: Kidney Renal Clear Cell Carcinoma; KIRP: Kidney Renal Papillary Cell Carcinoma; LAML: Acute Myeloid Leukemia; LGG: Brain Lower Grade Glioma; LIHC: Liver Hepatocellular Carcinoma; LUAD: Lung Adenocarcinoma; LUSC: Lung Squamous Cell Carcinoma; MESO: Mesothelioma; MF: Molecular Function; MHC: Major Histocompatibility Complex; MMR: Mismatch Repair; MSI: Microsatellites Instability; OS: Overall Survival; OV: Ovarian Serous Cystadenocarcinoma; PAAD: Pancreatic Adenocarcinoma; PCPG: Pheochromocytoma and Paraganglioma; PFS: Progression-Free Survival; PRAD: Prostate Adenocarcinoma; RCC: Renal Cell Carcinoma; READ: Rectum Adenocarcinoma; SARC: Sarcoma; SKCM: Skin Cutaneous Melanoma; STAD: Stomach Adenocarcinoma; TCGA: The Cancer Genome Atlas; TGCT: Testicular Germ Cell Tumors; THCA: Thyroid Carcinoma; THYM: Thymoma; TIL: Tumor-Infiltrating Lymphocytes; TIMER2.0: Tumor Immune Estimate Resource, Version 2; TMB: Tumor Mutation Burden; TME: Tumor Microenvironment; UCEC: Uterine Corpus Endometrial Carcinoma; UCS: Uterine Carcinosarcoma; WSTF: Williams Syndrome Transcription Factor
Introduction
Cancer occupies the forefront of the leading causes of death worldwide, endangering public health and quality of life [1]. In recent years, cancer immunotherapy, especially immune checkpoint blockade therapy, has come into focus [2]. Tumorigenesis is a complex process involving the immune system, and conventional therapeutic interventions are not satisfactory in cancer treatment. There is an urgent need for pan-cancer analysis of genes to assess clinical prognosis and the value of potential therapeutic targets. With the development and improvement of public databases such as The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO), we can perform pan-cancer expression, survival prognosis, and tumor immune infiltration assessment of genes in different tumors and analyze related signaling pathways [3,4].
The Bromodomain Adjacent to Zinc Finger Domain 1B (BAZ1B) gene encodes a member of the bromodomain family of proteins. The bromodomain is a structural motif characteristic of proteins involved in chromatin-dependent transcriptional regulation. This gene is missing in Williams-Beuren syndrome, a developmental disorder caused by the deletion of multiple genes in 7q11.23 [5]. BAZ1B has been reported to play a major role in DNA replication, DNA damage repair, RNA transcription, and apoptosis [6]. In addition, it is aberrantly expressed and dysregulated in many cancers. Currently, BAZ1B expression was found to be increased in breast [7], colon [8], and lung [9] cancers, which may predict a poor prognosis for patients. It was found that knockdown of BAZ1B or treatment of cells with BAZ1B antibody reduced cell proliferation and oncogenic pathway activation in colon cancer, suggesting that BAZ1B could be used as a cancer therapeutic target [8]. In addition, BAZ1B may promote lung cancer cells to undergo proliferation, promote colony formation, migration, and invasion by inducing epithelial mesenchymal transition and thus overexpression in lung cancer [9]. Although BAZ1B emerges as a promising cancer target, its expression, function and mechanism in various tumors remain unclear.
In this study, we performed a pan-cancer analysis of BAZ1B based on multiple databases. Factors such as gene expression and alterations, clinical prognosis, protein phosphorylation, immune infiltration and associated cellular pathways were addressed. Based on these results, we aimed to investigate the value as a biomarker and to assess the potential molecular mechanisms of BAZ1B in different cancers.
Materials and Methods
Gene expression analysis
In the “Gene” module (http://timer.comp-genomics.org/) of TIMER 2.0 (tumor immune estimate resource, version 2) [10], we observed the differential expression of BAZ1B in different TCGA tumors and adjacent normal tissues. For tumors where normal tissue was completely infiltrating or highly restricted, we used the module “Expression Analysis-Box Plots” of GEPIA2 (interactive analysis of gene expression profile, 2nd edition) (http:// gepia2. cancer-pku.cn) [11]. By adjusting the p value cut-off value = 0.01, |Log2FC| (fold change) cut-off value = 1, “matching TCGA normal and GTEX data”, we got the boxplots of the expression difference between the pathological tissues of these tumors and the corresponding normal tissues. In addition, the “Pathological staging map” module of GEPIA2 was used to analyze the violin plot expressed by BAZ1B in different stages of TCGA tumors. The expression data transformed by log2 [TPM (transcripts per million per) + 1] was applied to the violin graph. Notably, the UALCAN portal (http://ualcan.path.uab.edu/analysis-prot.html) [12] is an interactive portal that facilitates analysis of protein expression in CPTAC tumor datasets. We investigated the expression of total protein or phosphate protein of BAZ1B in primary tumor and normal tissues, respectively. The Human Protein Atlas (HPA) (https://www.proteinatlas.org/) is a database of cancer gene immunohistochemistry [13]. On the site, immunohistochemical techniques were used to show the distribution and expression of each protein in 48 normal human tissues and 20 tumor tissues. The protein expression of BAZ1B in urothelial cancer, colon adenocarcinoma, lymphoma, melanoma, testis germ cell tumor, endometrial cancer and corresponding normal human tissues was observed by immunohistochemical technique.
Genetic alteration and methylation analysis
We logged on to the website of Clinical Bioinformatics Assistant (https://www.aclbi.com), a database set containing 10228 cases involving 29 malignant tumors, to analyze BAZ1B's mutation analysis array in the "Pan-Cancer" module, including its Tumor Mutation Burden (TMB) and Microsatellites Instability (MSI). We applied the "Mutations" module of the cBioPortal database (https://www.cbioportal.org/) [14] to obtain mutation site information for BAZ1B and schematic diagrams of the three-dimensional structure. The "Comparison/Survival" module was used to explore the differences in Disease-Free Survival (DFS), Overall Survival (OS), Disease-Specific Survival (DSS), and Progression-Free Survival (PFS) rates of BAZ1B gene mutations in cancer patients, and we obtained Kaplan-Meier plots with log rank p values in cases of uterine corpus endometrial carcinoma
Protein phosphorylation analysis
The UniProt tool (https://www.uniprot.org/) was used to find the ID for the BAZ1B protein, and we then obtained the protein domain through the SMART dataset( http://smart.embl-heidelberg.de/ ). We obtained BAZ1B phosphorylation sites and protein expression levels in breast cancer and lung adenocarcinoma from the UALCAN portal.
Survival prognosis analysis
The Clinical Bioinformatics Assistant web was used to obtain overall, disease-free, disease-specific, and progression-free survival data maps of BAZ1B in TCGA tumors. The univariate cox regression analysis was carried out with R software v4.0.3, and the forest plot was used to show p value, Hazard Ratio (HR) and 95% Confidence Intervals (CI).
Immune infiltration analysis
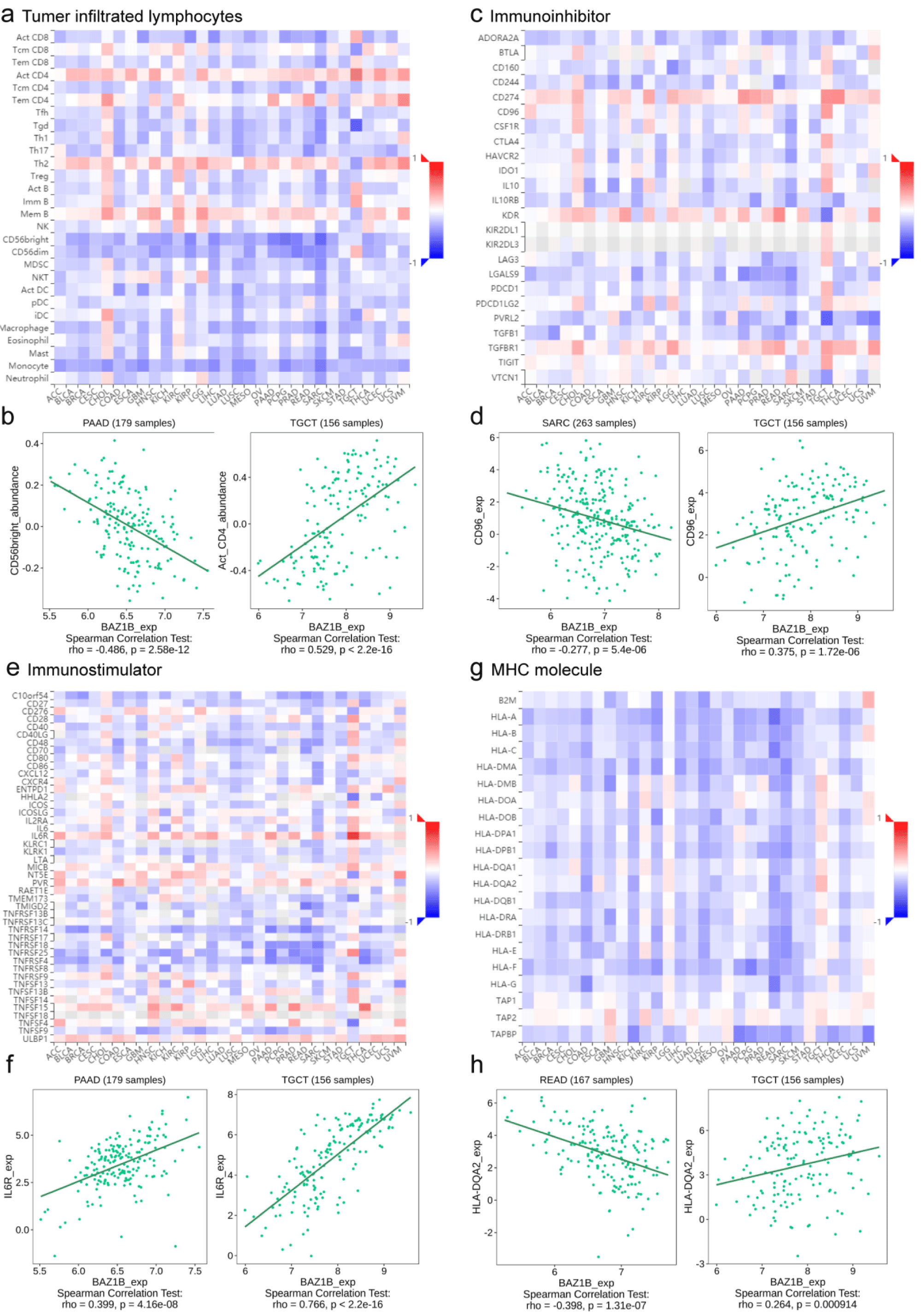
The TISIDB database (http://cis.hku.hk/TISIDB/index.php) [15] is an integrated repository portal for tumor-immune system interactions, which provides our assessment of the relationship between BAZ1B and immune traits including lymphocyte, immune modulators, and chemokines. We concluded the relationship between Tumor-Infiltrating Lymphocytes (TIL) and BAZ1B expression by searching the “Lymphocyte” section. Besides, we assessed the association of BAZ1B expression with immunoinhibitor, immunostimulator, and Major Histocompatibility Complex (MHC) molecule, respectively, using the “Immunomodulator” module.
BAZ1B-related gene enrichment analysis
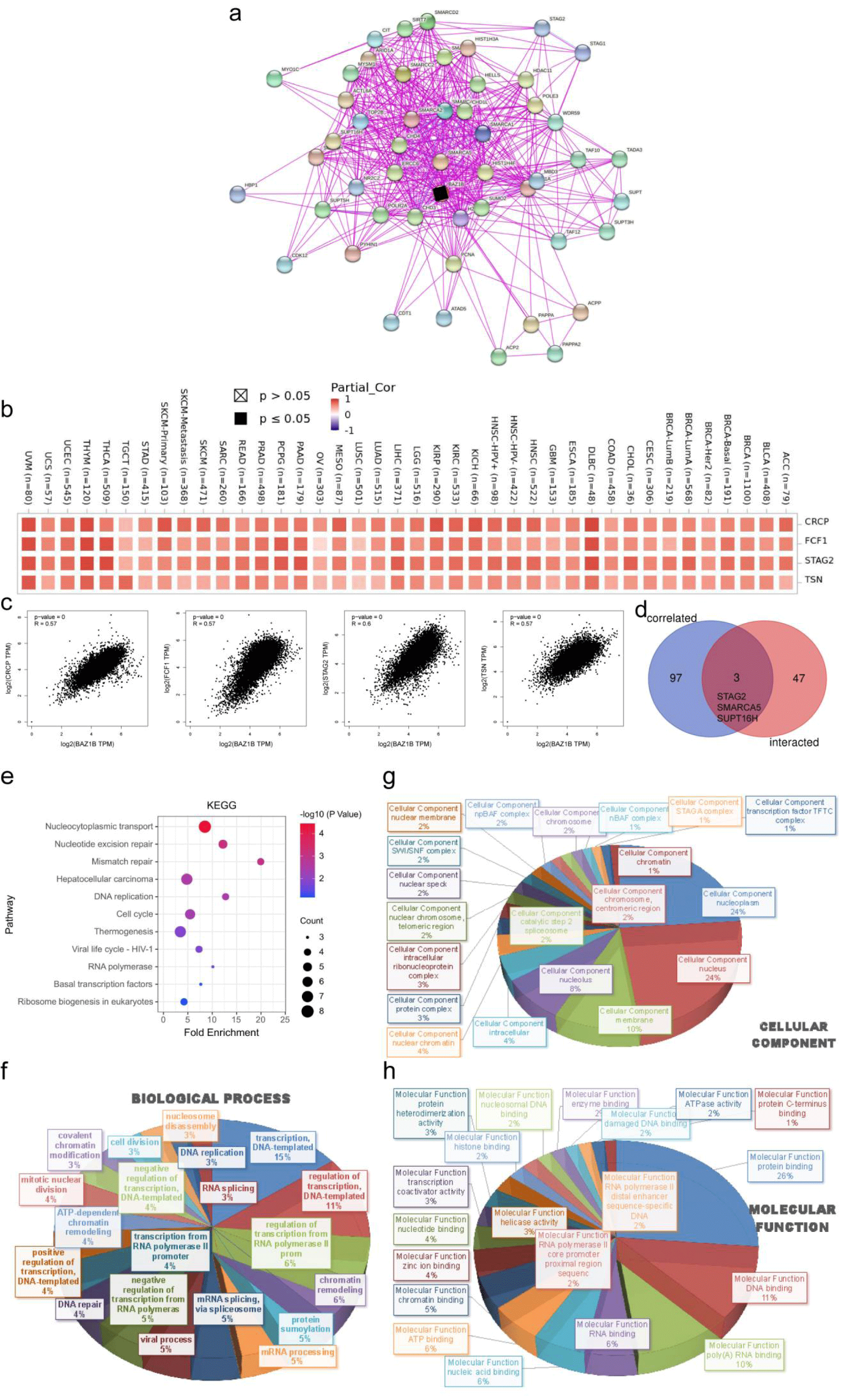
Based on the protein resource integration tool, STRING(https://string-db.org/) [16], we searched for “BAZ1B”and “Homo Sapiens” and chose a parameter setting: the minimum required interaction score ["Low confidence (0.150)"], the network edge meaning "evidence", the maximum number of interactors shown as "no more than 50 interactors" in 1st shell and the active interaction sources "experiments". The protein map associated with BAZ1B was generated by the screening. Under the “Similar Gene Detection” module of GEPIA2, 100 target genes was used in all TCGA tumor and normal tissue data sets. The “Correlation Analysis” model of paired Pearson correlation analysis identified whether BAZ1B was associated with established gene expression. Log2TPM, p value and correlation coefficient (R) were shown on the point graph. Furthermore, we used TIMER2’s “Gene_Corr” module to generate the heat map data for the selected genes, including the Spearman partial correlation and the purity adjusted P-level correlation test.
JVENN, an interactive venn graph viewer, was used to compare BAZ1B binding genes with interacting genes [17]. BAZ1B was analyzed for both the Kyoto Genome and Genome Encyclopedia (KEGG) pathways using two sets of data. We uploaded the gene list to DAVID (Database for Annotation, Visualization, and Integrated Discovery) (https://david.ncifcrf.gov/) [18] and set up the identifier and species as (“Official”) and “Homo Sapiens” to collect functional annotated map data. Then, the data were visualized as bubble plots via the HIPLOT website (https://hiplot.com.cn/). In addition, we imported the Biological Process (BP), Cellular Component (CC) and Molecular Function (MF) data into DAVID website and enriched them with Gene Ontology (GO) analysis. Finally, the Excel 2016 software was used to process the data and exported it into three-dimensional pie charts.
Statistical analysis
The Wilcox test was used to compare gene expression differences. Pearson correlation analysis was used to determine the relationship between BAZ1B and similar genes. Kaplan-Meier analysis, log-rank test and cox regression test were used in survival analysis. TIMER, CIBERSORT, CIBERSORT-ABS, QUANTISEQ, XCELL, MCPCOUNTER, and EPIC algorithms were used to estimate the immune penetration. By Spearman rank correlation test and purity adjustment method, p value and partial correlation value were obtained. In addition, the statistical significance was noted by the number of stars (*: p < 0.05; **: p < 0.01; ***: p < 0.001).
Results
Gene expression level
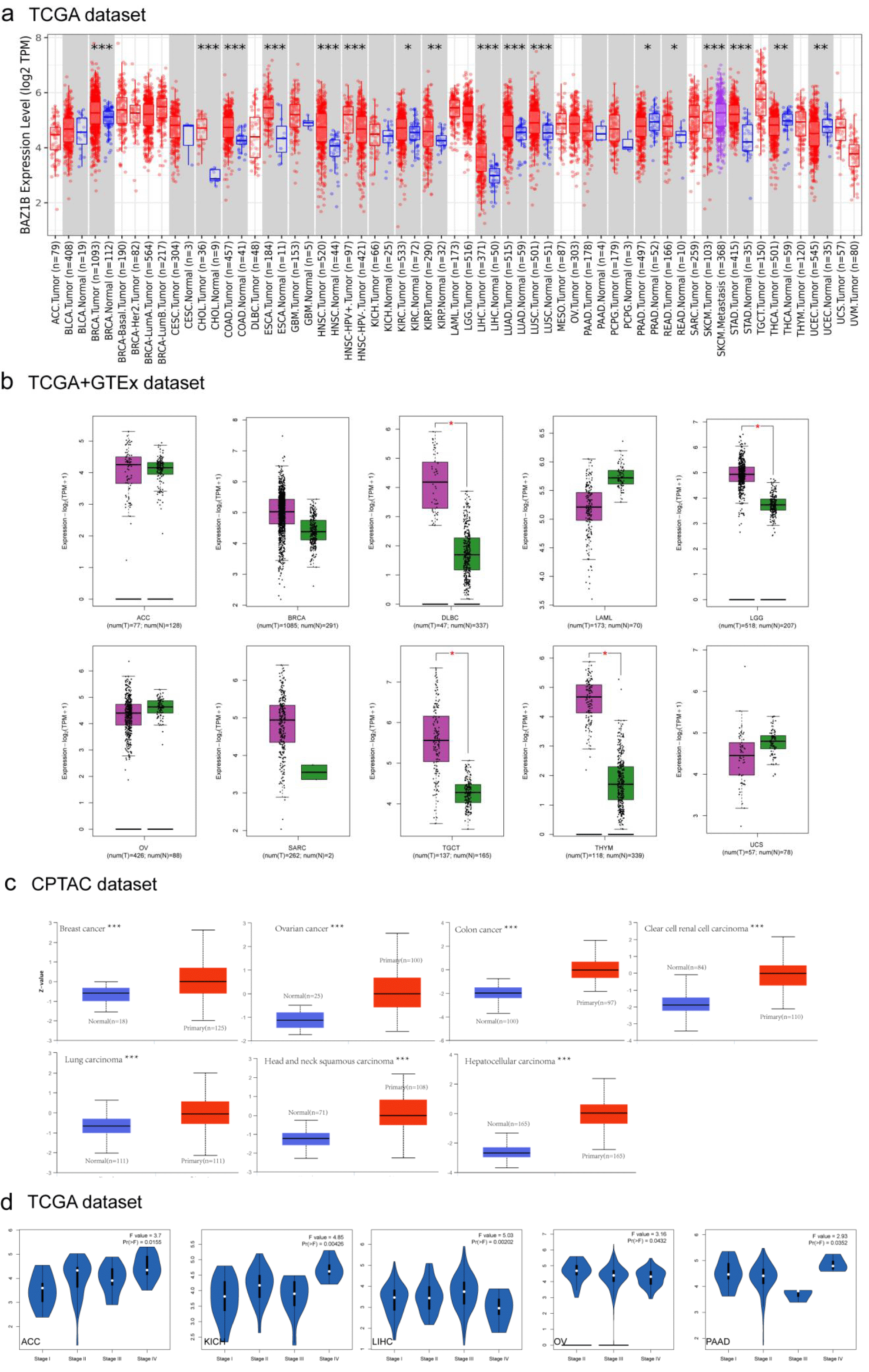
We analyzed the expression of BAZ1B in TCGA cancer by TIMER2.0. As shown in figure 1a, the BAZ1B expression level was higher in a variety of tumor tissues than in normal tissues, these included BRCA (Breast invasive carcinoma), CHOL (Cholangiocarcinoma), COAD (Colon Adenocarcinoma), ESCA (Esophageal Carcinoma), HNSC (Head and Neck Squamous Cell Carcinoma), LIHC (Liver Hepatocellular Carcinoma), LUAD (Lung Adenocarcinoma), LUSC (Lung Squamous Cell Carcinoma), SKCM (Skin Cutaneous Melanoma), STAD (Stomach Adenocarcinoma) (p < 0.001), KIRP (Kidney Renal Papillary Cell Carcinoma), THCA (Thyroid Crcinoma), UCEC (Uterine Corpus Endometrial Carcinoma) (p < 0.01) and KIRC (Kidney Renal Clear Cell Carcinoma), PRAD (Prostate Adenocarcinoma), READ (Rectum Adenocarcinoma) (p < 0.05). We further compared BAZ1B gene expression in some tumor and normal tissues in GEPIA2 with TCGA and GTEX databases (Figure 1b). In DLBC (Lymphoid neoplasm Diffuse Large B-Cell Lymphoma), TGCT (Testicular Germ Cell Tumors) and THYM (Thymoma), BAZ1B expression was significantly different from that in normal tissues (p < 0.05). However, there was no significant difference in ACC (Adrenocortical Carcinoma), OV (Ovarian Serous Cystadenocarcinoma), BRCA, LAML (Acute Myeloid Leukemia), SARC (Sarcoma) and UCS (Uterine Carcinosarcoma). Results from the CPTAC database showed that BAZ1B protein was expressed more frequently in BRCA, COAD, OV, KIRC, LUAD, HNSC and LIHC than in normal tissues (Figure 1c, p < 0.001). In order to get the expression level of BAZ1B in different tumor stages, we used the “Pathological Stage Plot” module in GEPIA2. BAZ1B gene expression in KICH, LIHC (p < 0.01), ACC, OV, PAAD (p < 0.05) was significantly different from pathological stage (Figure 1d).
Mutation analysis of BAZ1B gene in pan-cancer
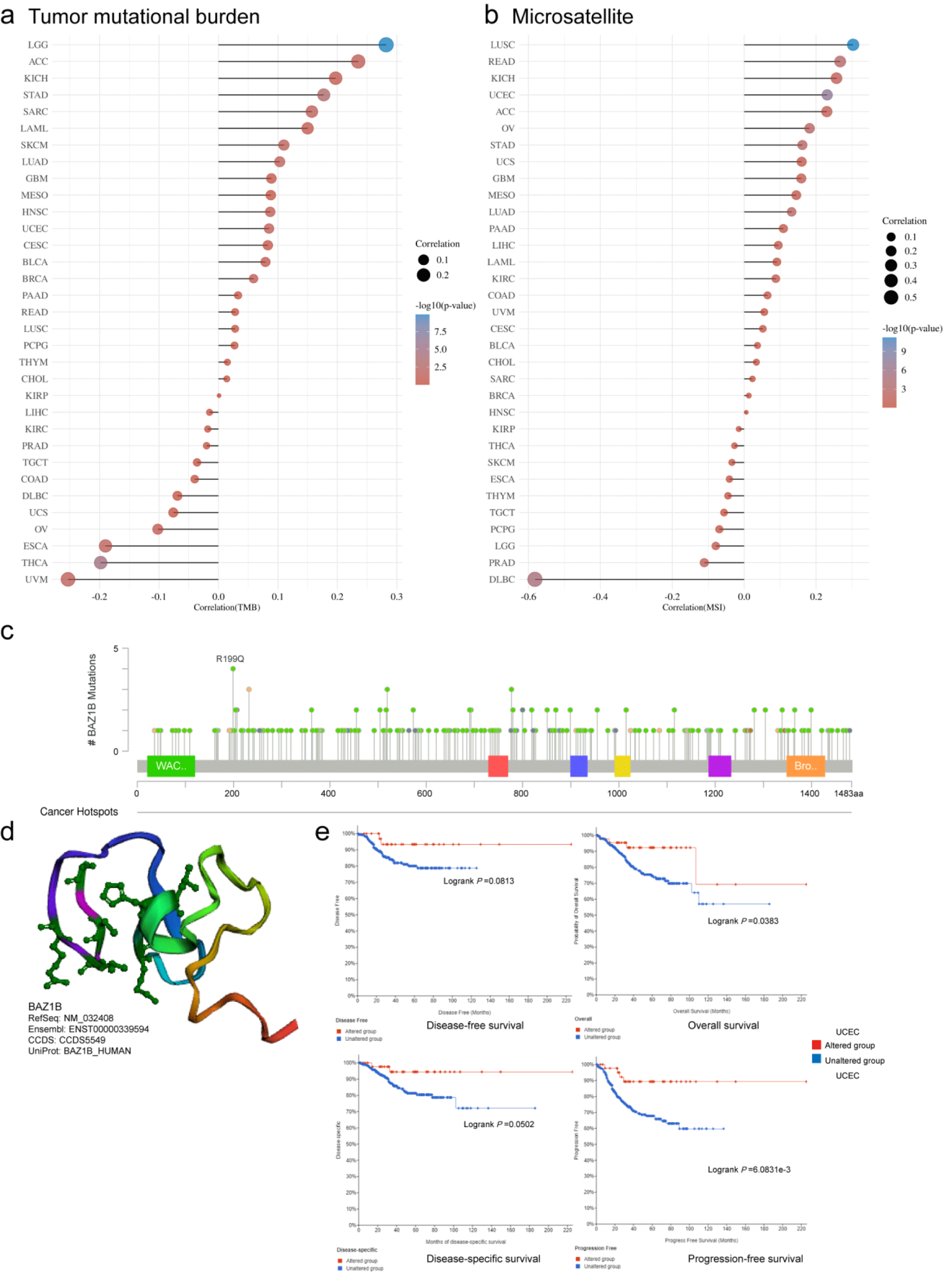
We analyzed the association of BAZ1B gene with TMB and MSI, respectively, in the Clinical Bioinformatics Assistant web (Figure 2a, 2b). In LGG (p < 0.001) and STAD (p < 0.01), BAZ1B was positively correlated with TMB, and BAZ1B was negatively correlated with TMB in UVM, ESCA (p < 0.05) and THCA (p < 0.01). In LUSC, UCEC (p < 0.001), READ (p < 0.01), BAZ1B was positively associated with MSI, but negatively associated with DLBC (p < 0.001). As shown in figure 2c, the type, site, and number of cases of genetic variation were clearly shown. We found that the missense mutation of BAZ1B was the main type of mutation, in which R199Q was changed in 1 case in UCEC and in 3 cases in COADREAD. We derived a 3D schematic of the structure of the BAZ1B protein (Figure 2d). In addition, we investigated the potential association between BAZ1B mutations and clinical survival in UCEC patients, patients with the BAZ1B alternations had poor disease-free survival (p = 0.0813), overall (p = 0.0383), disease specificity (p = 0.0502), and progression-free survival (p = 0.6831e-3) (Figure 2e).
Phosphorylation levels of BAZ1B protein in pan-cancer
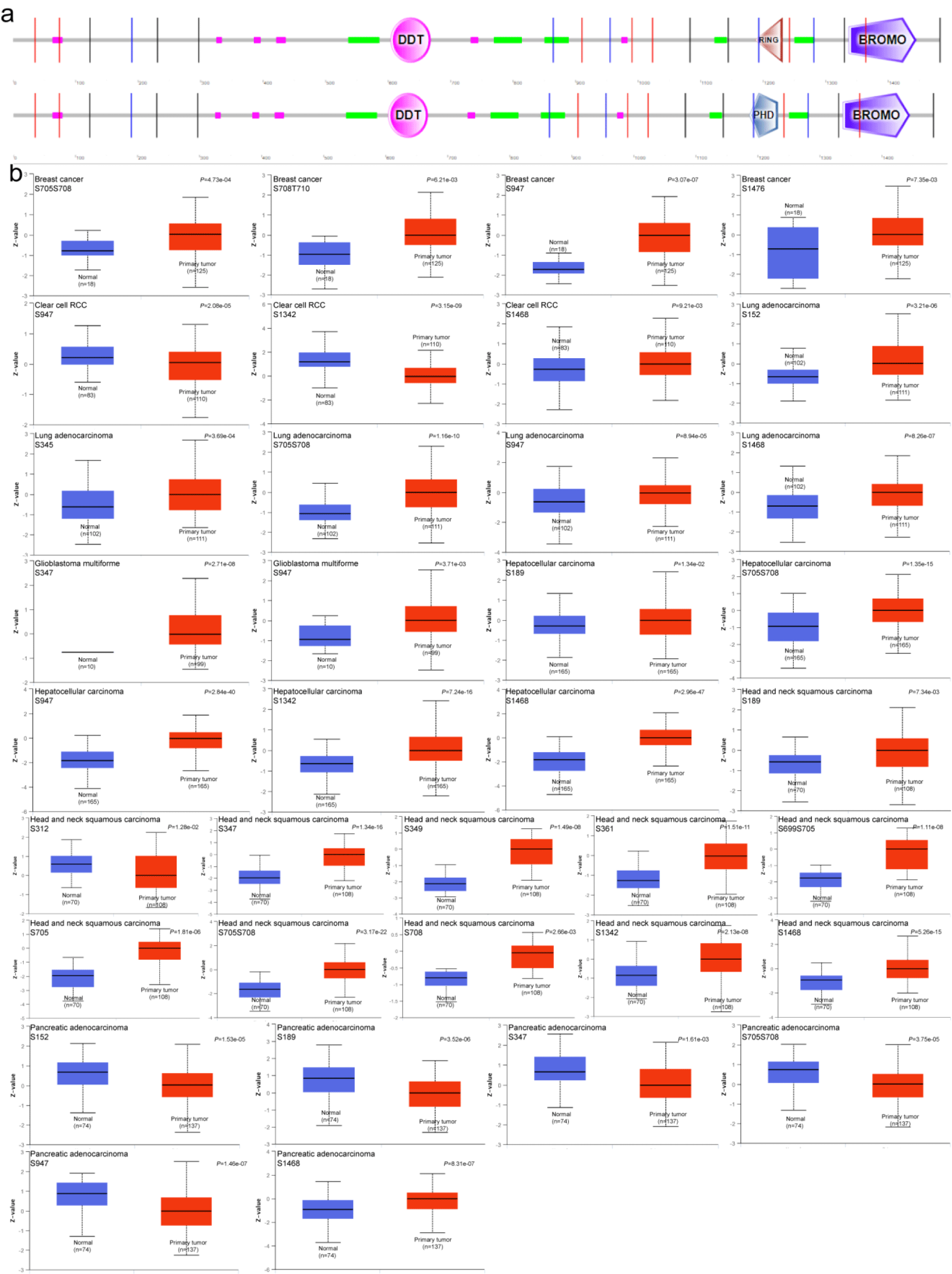
We summarized the graph of phosphorylation sites of the BAZ1B protein by retrieving the SMART tool (Figure 3a). Based on the CPTAC data set, we compared the levels of protein phosphorylation of BAZ1B between normal tissue and primary tumor. We analyzed seven types of tumors, including breast cancer, clear cells RCC, LUAD, HNSC, PAAD, GBM, and LIHC. The phosphorylation level of S947 site of BAZ1B in six primary tumor tissues was significantly different from that in normal tissues except for HNSC (p < 0.05). The phosphorylation levels of BAZ1B in five other cancers were higher than those in the control group, compared with HCC. Besides RCC and GBM, the phosphorylation level of S705S708 site of BAZ1B was also different in the other five primary tumors (p < 0.05), only in PAAD the phosphorylation level of this locus was lower than in the corresponding normal tissues. In addition, we observed that the S152 (p = 1.53e-05), S189 (p = 3.52e-06), S347 (p = 1.61e-03), S705S708 (p = 3.75e-05) and S1468 (p = 8.31e-07) of BAZ1B in PAAD showed decreased levels of phosphorylation compared with the control group. As shown in figure 3b, differences in the expression of other phosphorylation sites were analyzed and summarized. The results indicated that the phosphorylation level of BAZ1B in most primary tumors was significantly higher than that of the corresponding normal tissues.
Prognostic analysis of BAZ1B in pan-cancer
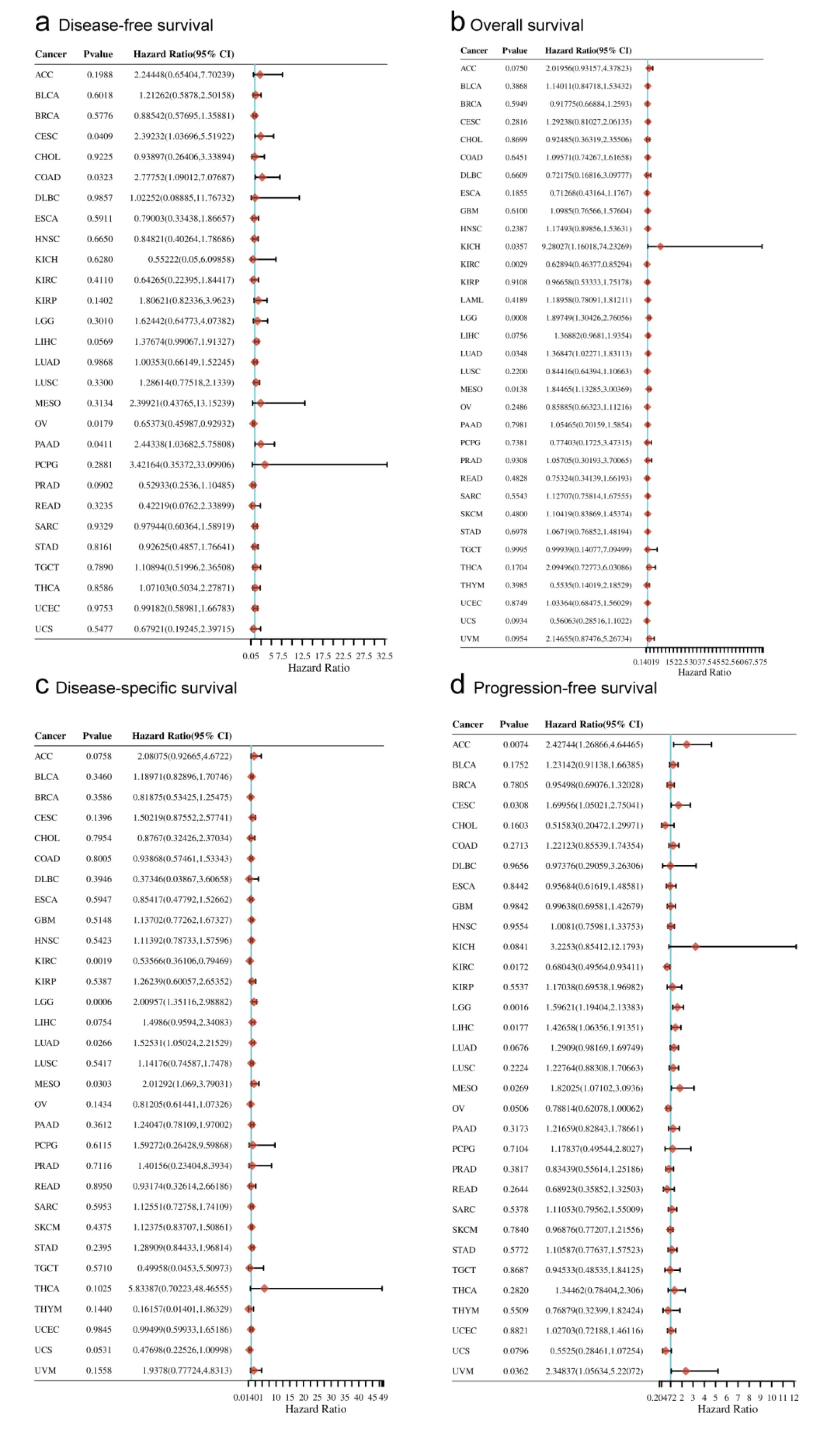
We summarized BAZ1B survival rates in 10,228 patients with 29 types of malignancy from the Clinical Bioinformatics Assistant website. High expression of BAZ1B was beneficial to the overall (p < 0.01), disease-specific, progression-free survival (both p < 0.05), and had no significant effect on disease-specific survival outcome in patients with KIRC compared with the corresponding normal tissues. Compared with the control group, the high expression of BAZ1B was lower associated with lower overall survival (p < 0.001), disease-specific, progression-free survival (both p < 0.01) in LGG patients and lower in MESO (Mesothelioma) (all p < 0.05), and there was no significant difference in disease-free survival among patients with cancer. In addition, we evaluated the prognostic value of BAZ1B in patients with other tumors, indicating that high BAZ1B expression may be a protective factor in KIRC, but in most other tumor patients, it implied a poor prognosis (Figures 4a-d).
Immunological value of BAZ1B in pan-cancer
TILs therapy, a kind of Adoptive Cell Therapy (ACT), is to isolate endogenous TILs from the lesion site of tumor patients through biopsy or surgery, then amplify them in vitro through Interleukin-2 (IL-2), and then infuse them back into patients. ACT and TILs have been shown to cause objective tumor regression in several types of cancer [19-23]. Therefore, it is necessary to assess the association of BAZ1B with the characteristics of pan cancer immune invasion. The results showed that BAZ1B was consistent with the expression levels of Th2 and Act_CD4 in most tumor patients, and was inversely correlated with most other types of TILs such as Act_CD8, CD56bright, etc. (Figure 5a). We observed that BAZ1B was opposite to CD56bright expression levels in PAAD (p = 2.58e-12) and consistent with Act_CD4 expression trends in TGCT (p < 2.2e-16). We also assessed the association of immunomodulators including immunoinhibitors, immunostimulators, MHC molecules and the expression of BAZ1B in pan cancer. As shown in figures 5b & 5c, most immunoinhibitors exhibited antagonism with BAZ1B in SARC patients, while the opposite was true in patients with TGCT. Figures 5d-f inferred the relationship between our randomly selected immunoinhibitor in SARC (p = 1.72e-06) and TGCT (p = 5.4e-06) and BAZ1B, respectively. MHC molecules are mediators on which T cells exert immune function when their surface is treated with antibodies to recognize antigens. When we observed the relationship between MHC molecules and BAZ1B in specific tumors from figure 5g, we found that in addition to TPA1 and TPA2, most MHC molecules antagonized the expression of BAZ1B in COAD, KICH, KIRP, READ, SARC. At the same time, we observed that there appears to be an exception in patients with TGCT tumors. Therefore, taking HLA-DQA2 as an example, we compared the differences in BAZ1B expression levels in READ (p = 1.31e-07) and TGCT (p < 0.001) (Figure 5h).
CD8+ T cells are the main immune cells that perform immune surveillance to detect antigens from cancer cells and developing malignant tumors [24]. Via Act_CD8+ T cells are reinfused into tumor patients, and BAZ1B overexpressed in most tumors may become a target in the process of destroying cancer cells by cell contact. CD96 in immune checkpoint inhibitor can regulate the antitumor effect of CD8+ T cells, and BAZ1B overexpressed in pan cancer was negatively correlated with it, which produced better therapeutic effect for patients with high TMB [25]. IL-6 is a factor that stimulates the proliferation, survival, invasion and metastasis of tumor cells, and also participates in the activation of T cells [26]. IL-6R can activate many signal pathways after binding cytokines [27]. Studies have shown that IL-6R is overexpressed in most tumors, and we also observed a positive correlation between BAZ1B and IL6R expression (Figure 5e). In summary, BAZ1B played an important role in tumor immunity.
Enrichment analysis of BAZ1B-related partners
We screened 50 genes that are strongly associated with the BAZ1B protein and supported by experimental evidence (Figure 6a). By interacting with 100 genes identified from GEPIA2 associated with BAZ1B expression, we obtained STAG2, SMARCA5, and SUPT16H. We used heat maps and scatter plots (Figures 6b-d, all p < 0.001) to show the correlation between the expression of the related genes CRCP, FCF1, STAG2, TSN and BAZ1B in pan-cancer. The results indicated that the related genes were positively associated with BAZ1B in most tumors. In addition, we analyzed the functional enrichment of their molecular mechanisms. Bubble graphs from KEGG data suggested that the “nucleocytoplasmic transport” and “nucleus excision repair” pathways may play a significant role in the tumorigenesis of BAZ1B (Figure 6e). The results of BP (Figure 6f), CC (Figure 6g) and MF (Figure 6h) in GO enrichment analysis data showed that most genes were related to the cell biology of DNA.
Conclusion
Pan-cancer analysis of any gene or genome that may be involved in cancer development, based on all public database platforms, will facilitate the refinement of malignant tumor intervention pathways and the design of immunological targets [28-30]. Given the complexity of cancer etiology and unsatisfactory treatment outcomes, it is imperative to explore the role of more genes in pan-cancer. BAZ1B has been reported to maintain chromosomal structural stability, participate in neurodevelopment, and be involved in cellular biological processes such as chromosomal remodeling, transcriptional regulation, DNA repair [31-35]. The literature has reported that high expression of BAZ1B can induce the development of a variety of cancers and is associated with poor prognosis. The BAZ1B gene plays a pro-tumor role in BRCA, COAD, and LUAC, and induces increased proliferation, migration, and invasion of cancer cells [7-9]. Based on public data platforms such as TCGA, CPTAC, and GEO, we detected the characteristics of gene expression, gene mutation, protein phosphorylation, immunological function, prognosis, and molecular mechanism of BAZ1B in pan-cancer.
MSI is a molecular indicator of DNA Mismatch Repair (MMR) deficiency and a biomarker of Immune Checkpoint Inhibitor (ICPI) response, and the high TMB and neoantigen load in MSI tumors favors immune effector cell infiltration and a stronger anti-tumor immune response within these tumors [36]. Over the past few years, MSI has become a major predictor of immune checkpoint blocking effects. We explored TMB and MSI levels of BAZ1B in pan-cancer. At the same time, we obtained the mutation site information of BAZ1B, and learned that the missense mutation occurring at the R199Q site made the prognosis of UCEC patients poor. We also observed phosphorylation site information for the BAZ1B protein in pan cancer, and interestingly, all of these showed statistical differences compared to the corresponding normal group.
The Williams Syndrome Transcription Factor (WSTF) is a hemizygous deletion gene encoded by the BAZ1B gene that has been identified as a hemizygous deletion gene in patients with Williams’s syndrome [37]. In addition to the functions of tyrosine protein kinase, WSTF protein also has the functions of BAZ1B's participation in transcription, replication, chromatin remodeling, and DNA damage response, which allow it to participate in a variety of cellular processes, including tumor cell proliferation and migration. We found abnormal expression of BAZ1B in a variety of tumors, including BRCA, CHOL, COAD, ESCA, HNSC, KIRC, LIHC, LUAD, LUSC, OV, and STAD. However, the prognostic form of BAZ1B brought to patients with different tumors varies. Survival analysis suggested that overexpression of BAZ1B was associated with poor prognosis in most tumors, especially in KIRC, LGG, LUAD, and MESO. This is consistent with the conclusion that Meng, et al. [9] in studying LUAD and Yang, et al. [38] in studying GBM, which both conclude that BAZ1B as an oncoprotein accelerates the proliferation and invasion of cancer cells among cancer patients. The downregulation of WSTF inhibits the proliferation, migration and invasion of cervical cancer cells [39] and may also be a new therapeutic target for estrogen-dependent breast cancer cell growth [40]. Taken together, these all mean that the high expression level of BAZ1B is related to the poor prognosis of tumor patients. In addition, our study analyzed other prognostic models, including PFS, DFS, and DSS.
Immune cells in TME have been reportedly associated with the proliferation and progression of cancer cells, thereby promoting or inhibiting tumors. Our findings were the first to suggest that BAZ1B expression was significantly associated with TIL, immunomodulators, and cancer-associated fibroblasts in pan-cancer, while implicitly reflecting immune status beyond prognosis. In addition, we integrated and analyzed the information of BAZ1B binding genes and BAZ1B-related partners, and obtained three common components STAG2, SMARCA5 and SUPT16H after cross-action. Our functional enrichment of BAZ1B for BAZ1B suggested that pathways such as changes in DNA cell biology may potentially lead to molecular mechanisms of cancer, which coincided with the findings of Sharif, et al. [6].
In summary, the first pan-cancer analysis of BAZ1B indicated a statistical correlation between BAZ1B expression and protein phosphorylation, survival prognosis, and tumor immune microenvironment in multiple tumors. Because BAZ1B was overexpressed in many cancers and accompanied by a poor prognosis, this increased the chance of becoming a new biomarker and therapeutic target. Our analysis also provided possible potential molecular mechanisms of BAZ1B, which better explain the role of BAZ1B in tumorigenesis from the perspective of clinical tumor samples.
References
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021 May;71(3):209-249. doi: 10.3322/caac.21660. Epub 2021 Feb 4. PMID: 33538338.
- Ribas A, Wolchok JD. Cancer immunotherapy using checkpoint blockade. Science. 2018 Mar 23;359(6382):1350-1355. doi: 10.1126/science.aar4060. Epub 2018 Mar 22. PMID: 29567705; PMCID: PMC7391259.
- Blum A, Wang P, Zenklusen JC. SnapShot: TCGA-Analyzed Tumors. Cell. 2018 Apr 5;173(2):530. doi: 10.1016/j.cell.2018.03.059. PMID: 29625059.
- Clough E, Barrett T. The Gene Expression Omnibus Database. Methods Mol Biol. 2016;1418:93-110. doi: 10.1007/978-1-4939-3578-9_5. PMID: 27008011; PMCID: PMC4944384.
- Peoples RJ, Cisco MJ, Kaplan P, Francke U. Identification of the WBSCR9 gene, encoding a novel transcriptional regulator, in the Williams-Beuren syndrome deletion at 7q11.23. Cytogenet Cell Genet. 1998;82(3-4):238-46. doi: 10.1159/000015110. PMID: 9858827.
- Sharif SB, Zamani N, Chadwick BP. BAZ1B the Protean Protein. Genes (Basel). 2021 Sep 28;12(10):1541. doi: 10.3390/genes12101541. PMID: 34680936; PMCID: PMC8536118.
- Lundqvist J, Kirkegaard T, Laenkholm AV, Duun-Henriksen AK, Bak M, Feldman D, Lykkesfeldt AE. Williams syndrome transcription factor (WSTF) acts as an activator of estrogen receptor signaling in breast cancer cells and the effect can be abrogated by 1α,25-dihydroxyvitamin D3. J Steroid Biochem Mol Biol. 2018 Mar;177:171-178. doi: 10.1016/j.jsbmb.2017.06.003. Epub 2017 Jun 10. PMID: 28610873.
- Liu Y, Wang SQ, Long YH, Chen S, Li YF, Zhang JH. KRASG12 mutant induces the release of the WSTF/NRG3 complex, and contributes to an oncogenic paracrine signaling pathway. Oncotarget. 2016 Aug 16;7(33):53153-53164. doi: 10.18632/oncotarget.10625. PMID: 27449290; PMCID: PMC5288175.
- Meng J, Zhang XT, Liu XL, Fan L, Li C, Sun Y, Liang XH, Wang JB, Mei QB, Zhang F, Zhang T. WSTF promotes proliferation and invasion of lung cancer cells by inducing EMT via PI3K/Akt and IL-6/STAT3 signaling pathways. Cell Signal. 2016 Nov;28(11):1673-82. doi: 10.1016/j.cellsig.2016.07.008. Epub 2016 Jul 21. PMID: 27449264.
- Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q, Li B, Liu XS. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020 Jul 2;48(W1):W509-W514. doi: 10.1093/nar/gkaa407. PMID: 32442275; PMCID: PMC7319575.
- Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019 Jul 2;47(W1):W556-W560. doi: 10.1093/nar/gkz430. PMID: 31114875; PMCID: PMC6602440.
- Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK, Varambally S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia. 2017 Aug;19(8):649-658. doi: 10.1016/j.neo.2017.05.002. Epub 2017 Jul 18. PMID: 28732212; PMCID: PMC5516091.
- Thul PJ, Lindskog C. The human protein atlas: A spatial map of the human proteome. Protein Sci. 2018 Jan;27(1):233-244. doi: 10.1002/pro.3307. Epub 2017 Oct 10. PMID: 28940711; PMCID: PMC5734309.
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012 May;2(5):401-4. doi: 10.1158/2159-8290.CD-12-0095. Erratum in: Cancer Discov. 2012 Oct;2(10):960. PMID: 22588877; PMCID: PMC3956037.
- Ru B, Wong CN, Tong Y, Zhong JY, Zhong SSW, Wu WC, Chu KC, Wong CY, Lau CY, Chen I, Chan NW, Zhang J. TISIDB: an integrated repository portal for tumor-immune system interactions. Bioinformatics. 2019 Oct 15;35(20):4200-4202. doi: 10.1093/bioinformatics/btz210. PMID: 30903160.
- Szklarczyk D, Gable AL, Nastou KC, Lyon D, Kirsch R, Pyysalo S, Doncheva NT, Legeay M, Fang T, Bork P, Jensen LJ, von Mering C. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021 Jan 8;49(D1):D605-D612. doi: 10.1093/nar/gkaa1074. Erratum in: Nucleic Acids Res. 2021 Oct 11;49(18):10800. PMID: 33237311; PMCID: PMC7779004.
- Bardou P, Mariette J, Escudié F, Djemiel C, Klopp C. jvenn: an interactive Venn diagram viewer. BMC Bioinformatics. 2014 Aug 29;15(1):293. doi: 10.1186/1471-2105-15-293. PMID: 25176396; PMCID: PMC4261873.
- Dennis G Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4(5):P3. Epub 2003 Apr 3. PMID: 12734009.
- Kumar A, Watkins R, Vilgelm AE. Cell Therapy With TILs: Training and Taming T Cells to Fight Cancer. Front Immunol. 2021 Jun 1;12:690499. doi: 10.3389/fimmu.2021.690499. PMID: 34140957; PMCID: PMC8204054.
- Stevanović S, Draper LM, Langhan MM, Campbell TE, Kwong ML, Wunderlich JR, Dudley ME, Yang JC, Sherry RM, Kammula US, Restifo NP, Rosenberg SA, Hinrichs CS. Complete regression of metastatic cervical cancer after treatment with human papillomavirus-targeted tumor-infiltrating T cells. J Clin Oncol. 2015 May 10;33(14):1543-50. doi: 10.1200/JCO.2014.58.9093. Epub 2015 Mar 30. PMID: 25823737; PMCID: PMC4417725.
- Li J, Chen QY, He J, Li ZL, Tang XF, Chen SP, Xie CM, Li YQ, Huang LX, Ye SB, Ke M, Tang LQ, Liu H, Zhang L, Guo SS, Xia JC, Zhang XS, Zheng LM, Guo X, Qian CN, Mai HQ, Zeng YX. Phase I trial of adoptively transferred tumor-infiltrating lymphocyte immunotherapy following concurrent chemoradiotherapy in patients with locoregionally advanced nasopharyngeal carcinoma. Oncoimmunology. 2015 Mar 6;4(2):e976507. doi: 10.4161/23723556.2014.976507. PMID: 25949875; PMCID: PMC4404907.
- Pedersen M, Westergaard MCW, Milne K, Nielsen M, Borch TH, Poulsen LG, Hendel HW, Kennedy M, Briggs G, Ledoux S, Nøttrup TJ, Andersen P, Hasselager T, Met Ö, Nelson BH, Donia M, Svane IM. Adoptive cell therapy with tumor-infiltrating lymphocytes in patients with metastatic ovarian cancer: a pilot study. Oncoimmunology. 2018 Sep 26;7(12):e1502905. doi: 10.1080/2162402X.2018.1502905. PMID: 30524900; PMCID: PMC6279323.
- Besser MJ, Shapira-Frommer R, Schachter J. Tumor-Infiltrating Lymphocytes: Clinical Experience. Cancer J. 2015 Nov-Dec;21(6):465-9. doi: 10.1097/PPO.0000000000000154. PMID: 26588677.
- Jiang X, Xu J, Liu M, Xing H, Wang Z, Huang L, Mellor AL, Wang W, Wu S. Adoptive CD8+ T cell therapy against cancer:Challenges and opportunities. Cancer Lett. 2019 Oct 10;462:23-32. doi: 10.1016/j.canlet.2019.07.017. Epub 2019 Jul 26. PMID: 31356845.
- Mittal D, Lepletier A, Madore J, Aguilera AR, Stannard K, Blake SJ, Whitehall VLJ, Liu C, Bettington ML, Takeda K, Long GV, Scolyer RA, Lan R, Siemers N, Korman A, Teng MWL, Johnston RJ, Dougall WC, Smyth MJ. CD96 Is an Immune Checkpoint That Regulates CD8+ T-cell Antitumor Function. Cancer Immunol Res. 2019 Apr;7(4):559-571. doi: 10.1158/2326-6066.CIR-18-0637. Epub 2019 Mar 20. PMID: 30894377; PMCID: PMC6445751.
- Kampan NC, Xiang SD, McNally OM, Stephens AN, Quinn MA, Plebanski M. Immunotherapeutic Interleukin-6 or Interleukin-6 Receptor Blockade in Cancer: Challenges and Opportunities. Curr Med Chem. 2018;25(36):4785-4806. doi: 10.2174/0929867324666170712160621. PMID: 28707587.
- Taher MY, Davies DM, Maher J. The role of the interleukin (IL)-6/IL-6 receptor axis in cancer. Biochem Soc Trans. 2018 Dec 17;46(6):1449-1462. doi: 10.1042/BST20180136. Epub 2018 Nov 22. PMID: 30467123.
- ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium. Pan-cancer analysis of whole genomes. Nature. 2020 Feb;578(7793):82-93. doi: 10.1038/s41586-020-1969-6. Epub 2020 Feb 5. PMID: 32025007; PMCID: PMC7025898.
- Huntsman DG, Ladanyi M. The molecular pathology of cancer: from pan-genomics to post-genomics. J Pathol. 2018 Apr;244(5):509-511. doi: 10.1002/path.5057. PMID: 29436707.
- Srivastava S, Hanash S. Pan-Cancer Early Detection: Hype or Hope? Cancer Cell. 2020 Jul 13;38(1):23-24. doi: 10.1016/j.ccell.2020.05.021. Epub 2020 Jun 11. PMID: 32531269.
- Xiao A, Li H, Shechter D, Ahn S, Fabrizio L, Erdjument-Bromage H, et al. WSTF regulates the H2A.X DNA damage response via a novel tyrosine kinase activity. Nature. 2009;457(7225):57-62. doi: 10.1038/nature07668. PubMed PMID: 19092802.
- Zhou J, Zheng Y, Liang G, Xu X, Liu J, Chen S, Ge T, Wen P, Zhang Y, Liu X, Zhuang J, Wu Y, Chen J. Atypical deletion of Williams-Beuren syndrome reveals the mechanism of neurodevelopmental disorders. BMC Med Genomics. 2022 Apr 4;15(1):79. doi: 10.1186/s12920-022-01227-7. PMID: 35379245; PMCID: PMC8981662.
- Sun H, Martin JA, Werner CT, Wang ZJ, Damez-Werno DM, Scobie KN, Shao NY, Dias C, Rabkin J, Koo JW, Gancarz AM, Mouzon EA, Neve RL, Shen L, Dietz DM, Nestler EJ. BAZ1B in Nucleus Accumbens Regulates Reward-Related Behaviors in Response to Distinct Emotional Stimuli. J Neurosci. 2016 Apr 6;36(14):3954-61. doi: 10.1523/JNEUROSCI.3254-15.2016. PMID: 27053203; PMCID: PMC4821908.
- Barnett C, Krebs JE. WSTF does it all: a multifunctional protein in transcription, repair, and replication. Biochem Cell Biol. 2011 Feb;89(1):12-23. doi: 10.1139/O10-114. Erratum in: Biochem Cell Biol. 2015 Aug;93(4):421. PMID: 21326359; PMCID: PMC3251257.
- Oppikofer M, Sagolla M, Haley B, Zhang HM, Kummerfeld SK, Sudhamsu J, Flynn EM, Bai T, Zhang J, Ciferri C, Cochran AG. Non-canonical reader modules of BAZ1A promote recovery from DNA damage. Nat Commun. 2017 Oct 11;8(1):862. doi: 10.1038/s41467-017-00866-0. PMID: 29021563; PMCID: PMC5636791.
- Karamitopoulou E, Andreou A, Wenning AS, Gloor B, Perren A. High tumor mutational burden (TMB) identifies a microsatellite stable pancreatic cancer subset with prolonged survival and strong anti-tumor immunity. Eur J Cancer. 2022 May 2;169:64-73. doi: 10.1016/j.ejca.2022.03.033. Epub ahead of print. PMID: 35512587.
- Kozel BA, Barak B, Kim CA, Mervis CB, Osborne LR, Porter M, Pober BR. Williams syndrome. Nat Rev Dis Primers. 2021 Jun 17;7(1):42. doi: 10.1038/s41572-021-00276-z. PMID: 34140529.
- Yang L, Du C, Chen H, Diao Z. Downregulation of Williams syndrome transcription factor (WSTF) suppresses glioblastoma cell growth and invasion by inhibiting PI3K/AKT signal pathway. Eur J Histochem. 2021 Nov 17;65(4):3255. doi: 10.4081/ejh.2021.3255. PMID: 34784707; PMCID: PMC8611414.
- Jiang D, Ren C, Yang L, Li F, Yang X, Zheng Y, Ji X, Tian Y. Williams syndrome transcription factor promotes proliferation and invasion of cervical cancer cells by regulating PI3K/Akt signaling pathway. J Obstet Gynaecol Res. 2021 Jul;47(7):2433-2441. doi: 10.1111/jog.14813. Epub 2021 May 24. PMID: 34028125.
- Lundqvist J, Hansen SK, Lykkesfeldt AE. Vitamin D analog EB1089 inhibits aromatase expression by dissociation of comodulator WSTF from the CYP19A1 promoter-a new regulatory pathway for aromatase. Biochim Biophys Acta. 2013 Jan;1833(1):40-7. doi: 10.1016/j.bbamcr.2012.10.012. Epub 2012 Oct 18. PMID: 23085504.
Content Alerts
SignUp to our
Content alerts.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.