Biology Group . 2023 May 04;4(5):849-852. doi: 10.37871/jbres1742.
Characterisation of a Group I Intron in the 18S rDNA of Thecamoeba (Amoebozoa; Discosea)
Daniele Corsaro*
- Free-living amoebae
- Thecamoeba
- Group I intron
- Nuclear intron
- 18S rDNA
Abstract
During an investigation of amoeba rDNA sequences, a group I intron was identified in the 18S rDNA of Thecamoeba aesculea (Amoebozoa; Discosea) at position S956. Secondary structure of Thecamoeba intron and phylogenetic analysis suggest that it is affiliated with other S956 introns occurring in distantly related amoebae such as acrasid (Percolozoa; Heterolobosea) and myxogastrid (Amoebozoa; Mycetozoa) slime moulds. This is the first report of a nuclear intron occurring within the genus Thecamoeba.
Introduction
Group I introns are mobile genetic elements that spread horizontally and/or vertically within the genomes of various organisms. They are self-splicing RNAs (ribozymes) that can also encode homing endonucleases allowing them to move to the DNA level [1,2]. In eukaryotes, group I introns occur mainly in the plastid and mitochondrial genomes, while their presence in nuclear genes is restricted to those of the ribosomal RNA operon in a wide and scattered set of microbial eukaryotes [1-3]. Group I introns have little sequence similarity but fold into conserved structures usually formed of nine paired elements (P1 to P9) and various loops. The overall structure and sequence variation in the conserved core (P3 to P8) allows their subdivision into five classes IA to IE [4,5]. Most nuclear introns are of IC and IE types, and they are particularly studied in fungi, various algae or plasmodial slime moulds (myxogastrid mycetozoans) because of their high frequency and variety [6-9]. On the other hand, they seem to occur more rarely in other protistan groups.
By studying the free-living amoeba Acanthamoeba (Amoebozoa; Discosea, Centramoebia), nuclear IC introns were found in the 18S rDNA of strains belonging to four distinct genotypes: T3 (A. griffini), T4A (A. castellanii complex), T5 (A. lenticulata), and T15 (A. jacobsi) [10-14]. During our studies on Acanthamoeba introns, an investigation of the available rDNA sequences of amoebae that may contain introns was conducted, based on data from the literature and BLAST analyses. Herein is reported the identification and characterization of a previously unnoticed intron in the 18S rDNA of Thecamoeba aesculea (Amoebozoa; Discosea, Flabellinia), a terrestrial amoeba isolated from epiphytic mosses and bark of the European horse-chestnut, Aesculus hippocastanum (Angiospermae, Sapindales) [15].
Materials and Methods
The intron characterized here is in the 18S rDNA of Thecamoeba aesculea (CCAP 1583/12) (GenBank ID JN247436, mislabelled as a mitochondrial sequence) [15,16]. Its identification was made during a wide screening of the available sequences to specifically detect nuclear introns. Once the intron was delimited, its secondary structure was deduced manually by identifying the conserved elements, therefore using Mfold for the remaining portions of the molecule, as described previously [13]. The phylogenetic relationships of the intron were deduced by analysing the extended conserved core (P3 to P8, P1’ and proximal sites of P2) [17] of selected nuclear introns of IC and IE classes. The sequences were manually aligned according to the secondary structure of the introns and phylogenetic trees were built with Maximum Likelihood (ML) (GTR+Γ model; 1000 replicates), Neighbour Joining (NJ) (Jukes-Cantor model; 2000 replicates) and maximum parsimony (MP; 1000 replicates), as described previously [13,14,17].
To verify whether the intron could have influenced the 18S rDNA-based phylogeny of the line, ML analysis (GTR+Γ+I:4; 1000 replicates) was performed on complete and partial (>1300 bp) sequences of Thecamoebidae (Sappinia, Stenamoeba, Thecamoeba, and Thecochaos), and the intron-free sequence of T. aesculea, using Dermamoebidae (Dermamoeba algensis and Paradermamoeba levis) as outgroup, previously aligned with MAFFT and refined for ambiguous sites using BioEdit, as described previously [18]. The same was done in parallel but with the intron-containing sequence of Thecamoeba. Both datasets were also analysed after realignment with MUSCLE.
Results and Discussion
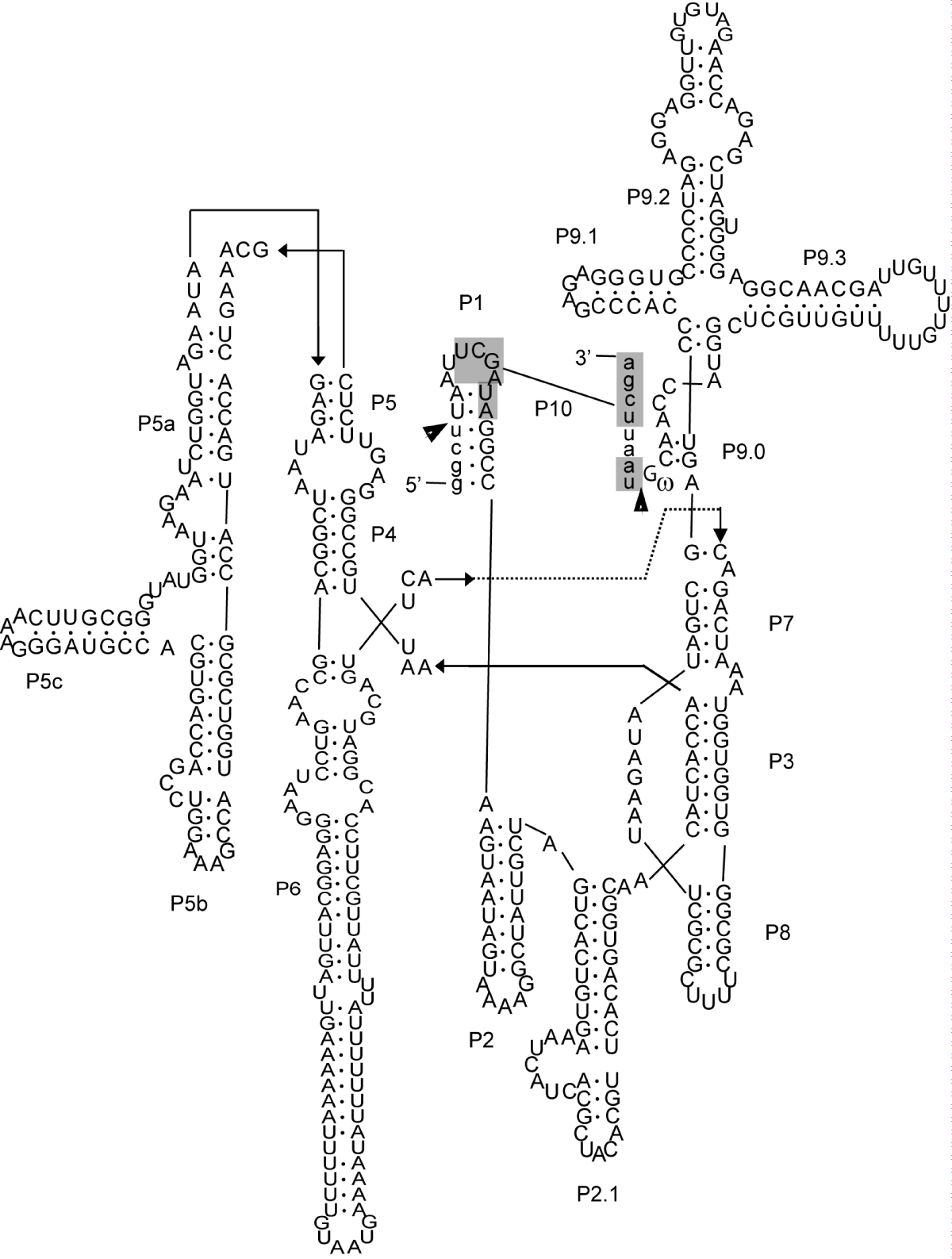
The 18S rDNA sequence of T. aesculea deposited in GenBank is 2224 nucleotides, a fairly usual length for Discosea whose members often have expansion regions in their 18S rDNA. The present analysis revealed that an intron of 432 nucleotides is actually inserted at position S956, in the loop between helices 34 and 35. As a result, the available rRNA of T. aesculea is only 1792 nt, ending with an incomplete H43-1 region. The presence of the Thecamoeba intron is not indicated in the GenBank file, nor is it reported in any publication on these amoebae. No introns were found in the more than 50 additional 18S rDNA sequences from Thecamoebidae analysed here.
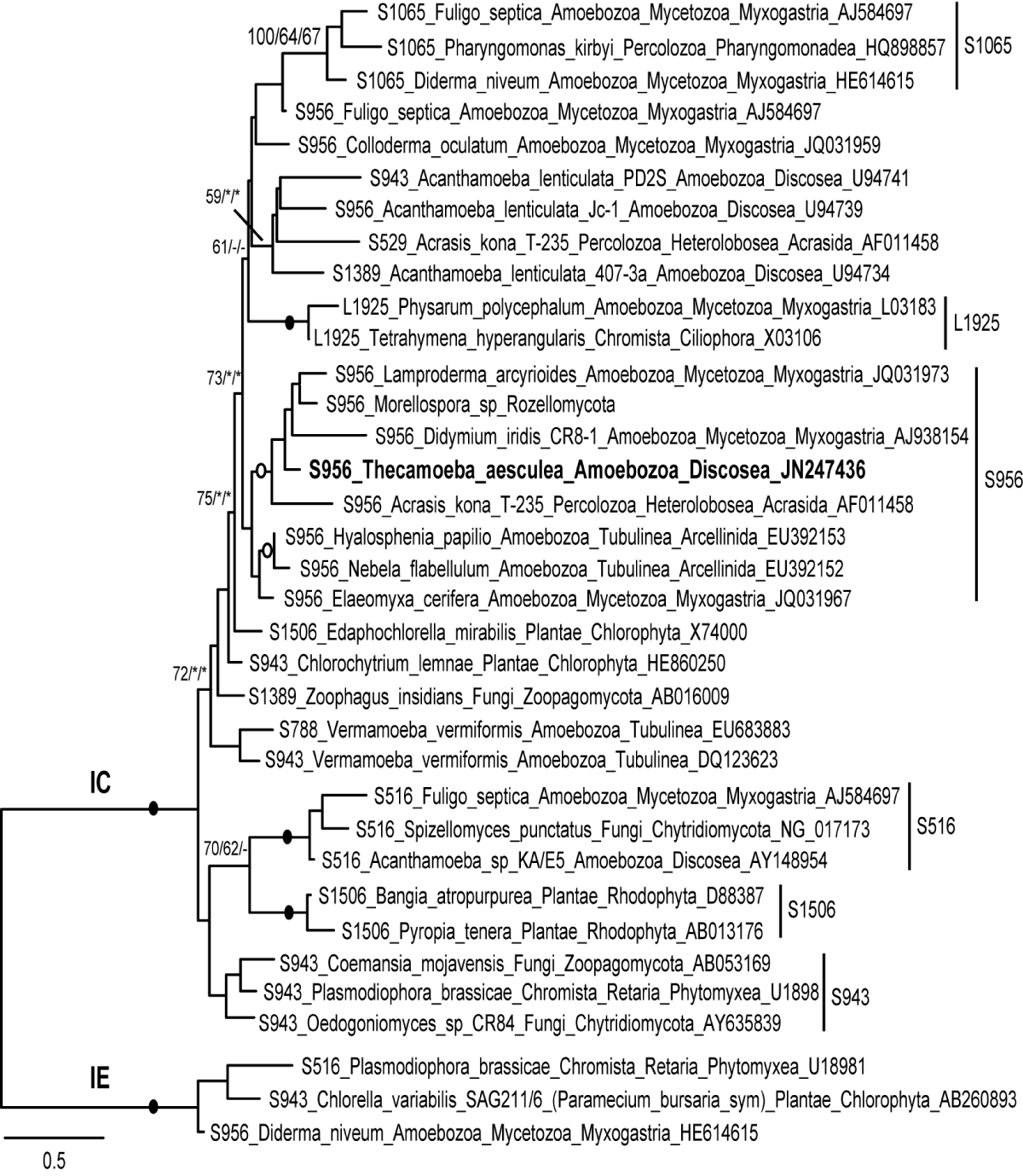
The secondary structure (Figure 1) and signature sequences indicate that the Thecamoeba intron belongs to the IC class. Phylogenetic analysis (Figure 2) suggests that it is close to other introns with the same insertion site, S956, occurring in distant organisms such as arcellinid testate amoebae (Hyaloshaenia, Nebela), Acrasis (Percolozoa; Heterolobosea, Acrasida), various myxogastrids (Amoebozoa; Mycetozoa) and Morellospora (Rozellomycota; Morellosporales). All have a similar secondary structure and size (~ 400 nt), except for the Didymium intron which encodes an endonuclease, and share 60 - 70% similarity with the Thecamoeba intron. It might be interesting to note that the intron-bearing Thecamoeba strain was isolated from tree bark [15], where acrasids and myxogastrids also live, and that Morellospora is a microsporidium-like intracellular parasite of amoebae, including slime moulds [19]. Close host-host interactions, such as those occurring during predation or parasitism, can facilitate lateral gene transfer. One could therefore suppose that these introns, belonging to the same lineage, can move horizontally between hosts sharing similar microhabitats and capable of interacting in various ways.
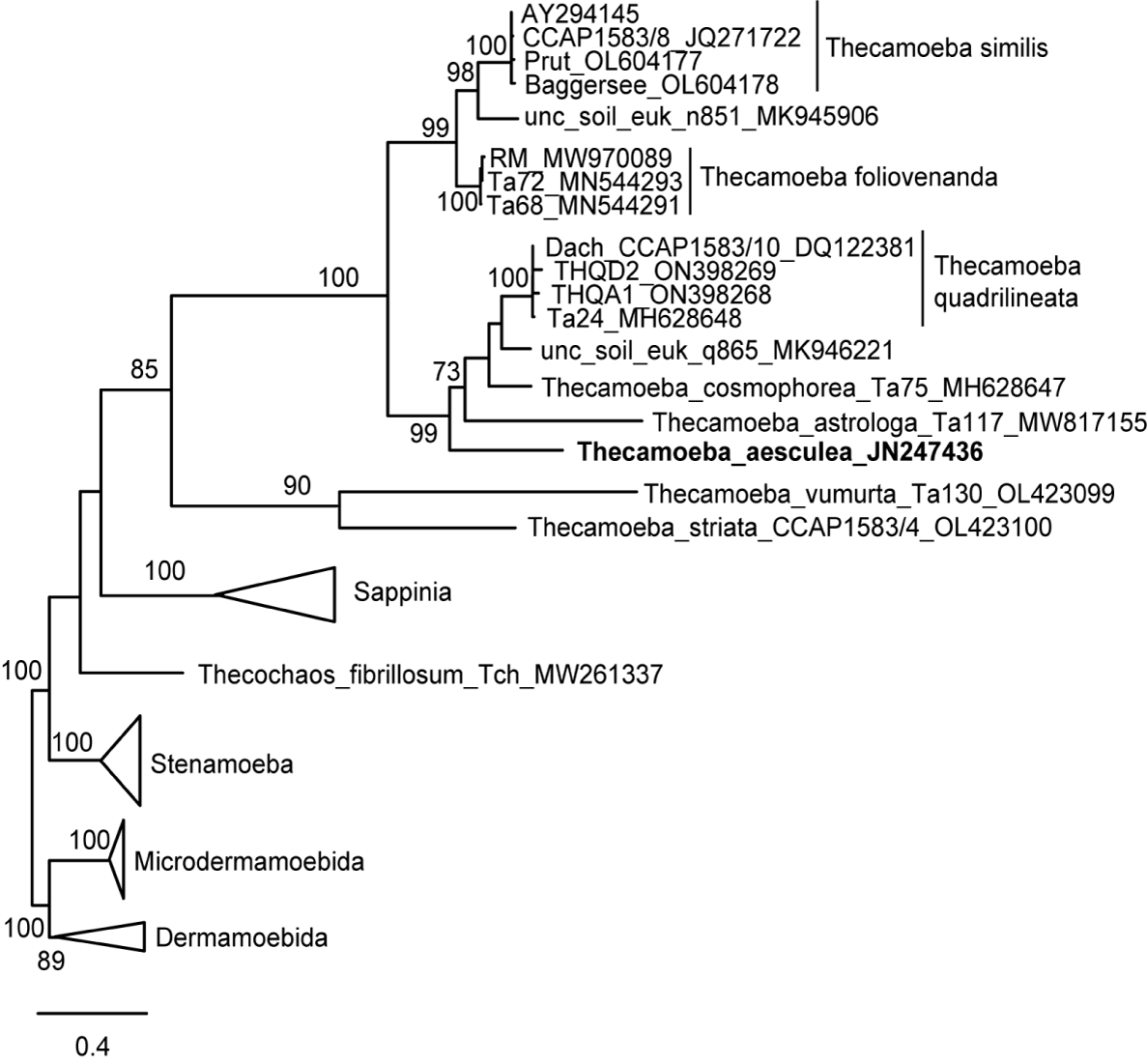
In 18S rDNA phylogeny based on intron-less sequences, T. aesculea emerges basal within the T. quadrilineata group (Figure 3), which also includes T. cosmophorea and T. astrologa, consistent with previous studies [20,21].
It should be noted that the presence of an intron in this sequence being missing, it could lead to misalignments. Introns insert at specific sites in conserved regions, normally causing large gaps in multiple alignments, making them easily recognizable. However, the same sequence may be treated differently by separate programs, and/or depending on the number and variety of other sequences analysed. In the special case of the Thecamoeba intron analysed here, it is noteworthy that the first eight nucleotides of the intron at its 5' end (P1 loop) are identical to the portion of the 18S rDNA located just after the S956 insertion site, on the 5' side of H35. In this way, the intron would not be recognized as a foreign element, instead appearing from its initial part as homologous to H35, thus inducing a misalignment, and its most divergent parts can be visualized as hypervariable regions or expansions of a few tens of not over the entire H34 to H44 portion of other Thecamoeba species. Such misalignment has indeed been produced in some cases, especially when few sequences from many distinct taxa are included. These regions will be removed during a phylogenetic analysis, thus having little effect on the result. Indeed, a virtually identical 18S tree topology can be obtained, differing for T. aesculea only in its branch length.
Conclusion
This is the first characterization of a group I intron present in the nuclear rDNA of a member of the genus Thecamoeba. Beyond Acanthamoeba spp., it is the only other member of the Discosea within which group I nuclear introns have been identified and characterized.
References
- Haugen P, Simon DM, Bhattacharya D. The natural history of group I introns. Trends Genet. 2005 Feb;21(2):111-9. doi: 10.1016/j.tig.2004.12.007. PMID: 15661357.
- Hedberg A, Johansen SD. Nuclear group I introns in self-splicing and beyond. Mob DNA. 2013 Jun 5;4(1):17. doi: 10.1186/1759-8753-4-17. PMID: 23738941; PMCID: PMC3679873.
- Corsaro D, Venditti D. Putative group I introns in the eukaryote nuclear internal transcribed spacers. Curr Genet. 2020 Apr;66(2):373-384. doi: 10.1007/s00294-019-01027-0. Epub 2019 Aug 28. PMID: 31463775.
- Michel F, Westhof E. Modelling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. J Mol Biol. 1990 Dec 5;216(3):585-610. doi: 10.1016/0022-2836(90)90386-Z. PMID: 2258934.
- Suh SO, Jones KG, Blackwell M. A Group I intron in the nuclear small subunit rRNA gene of Cryptendoxyla hypophloia, an ascomycetous fungus: evidence for a new major class of Group I introns. J Mol Evol. 1999 May;48(5):493-500. doi: 10.1007/pl00006493. PMID: 10198116.
- Bhattacarya D, Cannone JJ, Gutell RR. Group I intron lateral transfer between red and brown algal ribosomal RNA. Curr Genet. 2001 Aug;40(1):82-90. doi: 10.1007/s002940100227. PMID: 11570520.
- Simon D, Moline J, Helms G, Friedl T, Bhattacharya D. Divergent histories of rDNA group I introns in the lichen family Physciaceae. J Mol Evol. 2005 Apr;60(4):434-46. doi: 10.1007/s00239-004-0152-2. PMID: 15883879.
- Nandipati SC, Haugli K, Coucheron DH, Haskins EF, Johansen SD. Polyphyletic origin of the genus Physarum (Physarales, Myxomycetes) revealed by nuclear rDNA mini-chromosome analysis and group I intron synapomorphy. BMC Evol Biol. 2012 Aug 31;12:166. doi: 10.1186/1471-2148-12-166. PMID: 22938158; PMCID: PMC3511172.
- Furulund BMN, Karlsen BO, Babiak I, Haugen P, Johansen SD. Structural Organization of S516 Group I Introns in Myxomycetes. Genes (Basel). 2022 May 25;13(6):944. doi: 10.3390/genes13060944. PMID: 35741706; PMCID: PMC9223047.
- Gast RJ, Fuerst PA, Byers TJ. Discovery of group I introns in the nuclear small subunit ribosomal RNA genes of Acanthamoeba. Nucleic Acids Res. 1994 Feb 25;22(4):592-6. doi: 10.1093/nar/22.4.592. PMID: 8127708; PMCID: PMC307848.
- Schroeder-Diedrich JM, Fuerst PA, Byers TJ. Group-I introns with unusual sequences occur at three sites in nuclear 18S rRNA genes of Acanthamoeba lenticulata. Curr Genet. 1998 Jul;34(1):71-8. doi: 10.1007/s002940050368. PMID: 9683678.
- Corsaro D, Köhsler M, Montalbano Di Filippo M, Venditti D, Monno R, Di Cave D, Berrilli F, Walochnik J. Update on Acanthamoeba jacobsi genotype T15, including full-length 18S rDNA molecular phylogeny. Parasitol Res. 2017 Apr;116(4):1273-1284. doi: 10.1007/s00436-017-5406-1. Epub 2017 Feb 11. PMID: 28190156.
- Corsaro D, Venditti D. Nuclear Group I introns with homing endonuclease genes in Acanthamoeba genotype T4. Eur J Protistol. 2018 Oct;66:26-35. doi: 10.1016/j.ejop.2018.07.002. Epub 2018 Jul 18. PMID: 30071371.
- Corsaro D, Köhsler M, Venditti D, Rott MB, Walochnik J. Recovery of an Acanthamoeba strain with two group I introns in the nuclear 18S rRNA gene. Eur J Protistol. 2019 Apr;68:88-98. doi: 10.1016/j.ejop.2019.01.007. Epub 2019 Jan 28. PMID: 30743186.
- Kudryavtsev A, Hausmann K. Thecamoeba aesculea n. sp. (Amoebozoa, Thecamoebidae), a terrestrial amoeba with affinities to Th. sphaeronucleolus (Greeff. 1891). Acta Protozool. 2009 48(2):91-6.
- Smirnov AV, Chao E, Nassonova ES, Cavalier-Smith T. A revised classification of naked lobose amoebae (Amoebozoa: lobosa). Protist. 2011 Oct;162(4):545-70. doi: 10.1016/j.protis.2011.04.004. Epub 2011 Jul 28. PMID: 21798804.
- Corsaro D, Venditti D. Nuclear group I introns in the 18S rDNA subtypes of Vermamoeba vermiformis. Biologia. 2022 Jul ;77(7):1899-1907. doi: 10.1007/s11756-022-01047-1.
- Corsaro D, Walochnik J, Köhsler M, Rott MB. Acanthamoeba misidentification and multiple labels: redefining genotypes T16, T19, and T20 and proposal for Acanthamoeba micheli sp. nov. (genotype T19). Parasitol Res. 2015 Jul;114(7):2481-90. doi: 10.1007/s00436-015-4445-8. Epub 2015 Apr 15. PMID: 25869957.
- Corsaro D, Walochnik J, Venditti D, Hauröder B, Michel R. Solving an old enigma: Morellospora saccamoebae gen. nov., sp. nov. (Rozellomycota), a Sphaerita-like parasite of free-living amoebae. Parasitol Res. 2020 Mar;119(3):925-934. doi: 10.1007/s00436-020-06623-5. Epub 2020 Feb 11. PMID: 32048025.
- Mesentsev YS, Smirnov AV. Thecamoeba cosmophorea n. sp. (Amoebozoa, Discosea, Thecamoebida) - An example of sibling species within the genus Thecamoeba. Eur J Protistol. 2019 Feb;67:132-141. doi: 10.1016/j.ejop.2018.12.003. Epub 2018 Dec 18. PMID: 30616106.
- Mesentsev Y, Smirnov A. Thecamoeba astrologa n. sp. - A new species of the genus Thecamoeba (Amoebozoa, Discosea, Thecamoebida) with an unusually polymorphic nuclear structure. Eur J Protistol. 2021 Oct;81:125837. doi: 10.1016/j.ejop.2021.125837. Epub 2021 Aug 14. PMID: 34583223.
Content Alerts
SignUp to our
Content alerts.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.