Biology Group. 2023 October 11;4(10):1394-1404. doi: 10.37871/jbres1809.
Diversity of Non-Influenza Respiratory Viruses Associated with Influenza-Like Illness during 2009 pre and pandemic periods in São Paulo, Brazil, a Historical Overview
Renato de Sousa Paulino, Daniela Bernardes Borges da Silva, Katia Corrêa de Oliveira Santos, Margarete Aparecida Benega and Terezinha Maria de Paiva*
- Influenza-like illness
- Pandemic
- Respiratory viruses non-influenza
- Laboratory diagnosis
- Public health surveillance
- Disease prevention
Abstract
Background and objective: During the 2009 influenza A (H1N1) pandemic, 60% of the respiratory specimens collected from patients under surveillance for influenza-like illness (ILI) in São Paulo, Brazil, tested negative for influenza A and B by Real-Time RT-PCR (RT-qPCR) assays. In this retrospective study, we identified the diversity of other respiratory viruses associated with ILI during the pandemic period and added the benefit of RT-qPCR for their detection.
Methods: Five-hundred and forty-two influenza RT-qPCR negative respiratory specimens from ILI patients collected from Jan - Dec 2009, that also tested negative by a commercial Indirect Immuno Fluorescence (IIF) assay for the Respiratory Syncytial Virus (RSV), Parainfluenza (PIV) PIV1, PIV2, PIV 3 and Adenovirus (AdV) were retested by a panel of RT-qPCR assays for the other respiratory viruses, including RSV, Human Metapneumovirus (HMPV), PIV 1-4, AdV, Human Coronaviruses (HCoV) OC43, 229E, NL63 and HKU1, Rhinovirus (RV), Enterovirus (EV) and Human Bocavirus (HBoV).
Results: Of the 542 negative specimens, 428 (78%) were positive by RT-qPCR for another respiratory virus: In descending order: RV (149, 27%); RSV (41, 8%); AdV (32, 6%); HMPV (29, 5%); PIV3 (19, 4%); PIV1 (3, 1%); HCoV-OC43 (9, 2%); HBoV (9, 2%); PIV4 (6, 1%); HCoV-HKU1 (6, 1%); EV (5, 1%); HCoV-229E 4 (4, 1%); HCoV-NL63 (3, 1%). Co-detections of two viruses (98, 18%) and three viruses (12, 2%) were common.
Conclusion: This retrospective study revealed a high prevalence of respiratory viruses other than influenza associated with ILI during the pandemic period and highlights the superiority of RT-qPCR over IIF for ILI surveillance.
Introduction
Influenza-Like Illness (ILI), defined by the World Health Organization (WHO) as an acute respiratory infection with fever ≥ 38°C and symptoms onset within 10 days [1], is a leading cause of human morbidity and mortality worldwide [2]. As the name implies, influenza viruses are a predominant cause of ILI and consequently a major contributor to the human disease burden. However, ILI is a syndrome that is not specific to influenza virus and can be caused by many other respiratory pathogens as revealed by the increasing use of molecular diagnosistics [3]. In addition to the plethora of historically well-known respiratory viruses, molecular diagnostics has also contributed to the discovery of an increasing number of “new” respiratory viruses responsible for ILI in the last two decades, including Human Metapneumovirus (HMPV), 2001 [4]; Human Coronaviruses NL63 (HCoV-NL63), 2004 [5]; Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV), 2003 [6,7]; (HCoV-HKU1),2005 [8]; Human Bocavirus (HBoV), 2005 [9]; two new polyomaviruses WU (2007) [10] and KI (2007) [11]; new Human Rhinovirus (HRV - QPM), 2007 [12]; Middle East Respiratory Syndrome Coronavirus (MERS-CoV), 2012 [13].
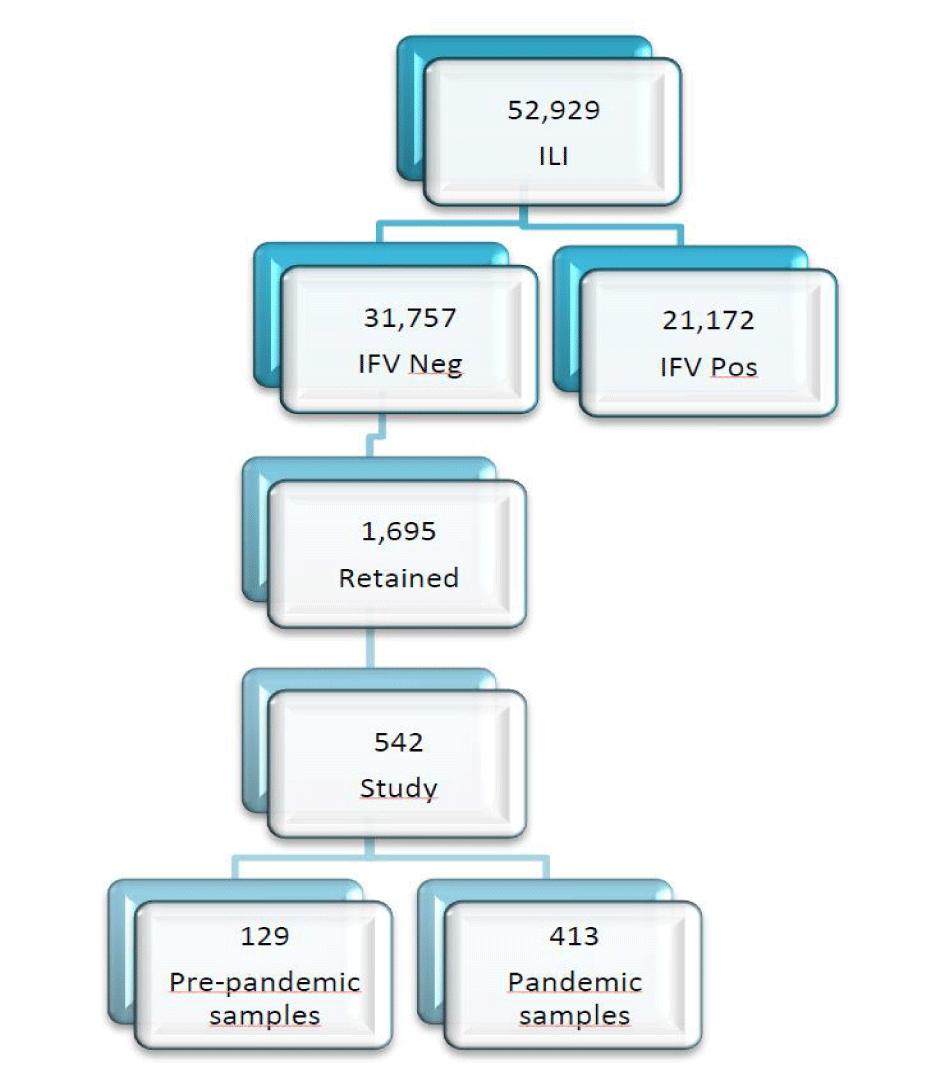
From January through December 2009, 52,929 respiratory specimens were collected from patients presenting with ILI, until five days after the clinical symptoms onset, were sent to the Respiratory Virus Laboratory (RVL)/Institute Adolfo Lutz (IAL), a National Influenza Centre/World Health Organization (NIC/WHO), to investigate an outbreak of a new pandemic strain of influenza virus, A(H1N1) [14] Sixty percent of these specimens tested negative for influenza A(H1N1), A(H1N1)pdm09, A(H3N2) and B viruses by real-time RT-qPCR assays (Figure 1). The aim of this retrospective study was to test a subset of these influenza-negative specimens using RT-qPCR assays to investigate the diversity of other respiratory viruses associated with ILI before and during the 2009 influenza pandemic.
Materials and Methods
Pre-study surveillance
IAL is an accredited NIC/WHO established in São Paulo, Brazil in 1960. In 2000, the Brazilian National Influenza Surveillance Network (NISN), sponsored by the Brazilian Ministry of Health, initiated surveillance for ILI in Brazil, and in 2002, IAL joined the NISN. Routine ILI surveillance consisted of receiving systematically collected Nasopharyngeal (NP) swab specimens from 5 children aged < 5 years old, per week, presenting with ILI symptoms at selected sentinel units: a children’s hospital, Hospital Municipal Infantil Menino Jesus; and two general hospitals, Municipal Ver José Storopolli and Geral de Guarulhos. Following the first confirmed case of influenza A (H1N1)pdm09 in São Paulo on May 07, 2009, ILI samples collection substantially increased from an expanded network of sentinel units at public and private hospitals located in São Paulo state and other Midwestern states: Mato Grosso (MT); Mato Grosso do Sul (MS), Goiânia (GO); Distrito Federal (DF); Northeast: Piauí (PI); and North: Acre (AC), Rondônia (RO); Tocantins (TO).
From January through December 2009, 52,929 respiratory specimens were collected at sentinel clinics from patients presenting with ILI and sent to the RVL//IAL/NIC/WHO for rapid processing and screening for influenza and other respiratory viruses Figure 1. All specimens were collected after the informed consent of parents or guardians following the Brazilian Ministry of Health protocol and with the approval of the IAL institutional review board.
After the first confirmed case of influenza pandemic strain A(H1N1)pdm09, in São Paulo (April 2009), from a respiratory secretion collected from an individual with a travel history to a risk area, the number of respiratory secretions collected from patients, presenting with Influenza-Like Illness (ILI) by the Children and General Sentinel Hospitals and public and private Hospitals, increased substantially. These clinical specimens were sent to the RVL/IAL/NIC/WHO for rapid processing and screening by Indirect Immunofluorescence (IIF) assays for other respiratory viruses (Figure 1). The clinical specimens corresponding to both pre- (129) and pandemic (413) periods were also tested by real-time (RT- qPCR) assays developed by the Centers for Disease Control and Prevention (CDC), Atlanta (USA), for the new influenza A(H1N1)pdm09 strain [15], seasonal influenza strains A(H1N1, H3N2)) and influenza B viruses were also tested by real-time RT - PCR (RT-qPCR). All samples were maintained at -80°C at the RVL/IAL/NIC/WHO. Approximately 60% of these specimens were negative for influenza viruses, raising the question of what other respiratory viruses might be responsible for ILI during this period. To answer this question, we selected 542 specimens, among the 1695 negative samples, by epidemiological week from January – December 2009, to test by CDC real-time RT-PCR (RT-qPCR) assays for non-influenza respiratory viruses. Results were negative using differential diagnosis (Influenza A, Influenza B, Respiratory Syncytial Virus (RSV), Parainfluenza Virus (PIVs) PIV-1, PIV-2, PIV3 and Adenovirus (AdV) by IIF, using Respiratory Panel 1 Viral Screening and Identification kit (Light Diagnostics, Chemicon, Temecula, CA) catalog number 3105 and also negative for influenza viruses by real-time RT – PCR (RT-qPCR).
Pre-study laboratory procedures
Routine viral surveillance was performed using commercial indirect immunofluorescence assays IIF assays for influenza viruses A and B, Respiratory Syncytial Virus (RSV), Parainfluenza (PIV1, PIV2, PIV3) and Adenovirus (AdV) available with the respiratory panel 1 viral screening and identification kit (Light Diagnostics, Chemicon, Temecula, CA) catalog number 3105. Assays were performed according to the manufacturer´s instructions.
Beginning on (29 May 2009), (RT-qPCR) assay for influenza A(H1N1), A(H1N1) pdm09, A(H3N2), and influenza virus B following procedures developed by the U.S. Centers for Disease Control and Prevention (CDC) were introduced [15].
Study laboratory procedures
Of the 52,929 surveillance specimens obtained 31,757 (60%) were negative for influenza A and B by the IIF panel and 1,695 of these were negative for influenza A and B by RT-qPCR assay and all other respiratory viruses covered by IIF panel Figure 1. These specimens were retained at -80C at the RVL/IAL/NIC/WHO.
Phases: In-house single plex real-time RT - PCR (RT-qPCR) assays for RSV, HMPV, PIV1 – PIV4, AdV, RV, enterovirus (EV), HBoV, and HCoVs 229E, OC43, NL63, and HKU1 following procedures developed by CDC. The primers and probes reflect the respiratory virus sequences obtained from the databases at the time. The CDC respiratory virus group likely had them updated. Real-Time RT-PCR (RT-qPCR) was conducted by using AgPath _ ID TM (Applied Biosystems) A positive test result was considered a well-defined fluorescence curve crossing the threshold within 40 cycles. All runs included positive and negative virus template controls to monitor assay performance. The Ct values of positive samples are shown in table 1.
| Table 1: In house assay Ct values, mean, range. In house assay results classified as strong (Ct < 30) moderate (Ct ≥ 30 to ≤ 37) or weak (Ct > 37 to < 40) positive. | ||||||
| Ct Range | ||||||
| Target | (n) | Mean Ct | SD | Min | Max | |
| RV | Samples | 195 | 29.29 | 4.49 | 15.94 | 37.71 |
| Control | 63 | 28.93 | 1.58 | 26.55 | 34.36 | |
| AdV | Samples | 74 | 31.67 | 5.93 | 13.75 | 39.48 |
| Control | 63 | 29.10 | 1.18 | 24.36 | 32.23 | |
| RSV | Samples | 63 | 27.84 | 6.21 | 17.44 | 38.72 |
| Control | 63 | 30.57 | 1.16 | 26.68 | 32.84 | |
| HMPV | Samples | 56 | 27.80 | 4.85 | 20.30 | 38.97 |
| Control | 63 | 30.06 | 0.87 | 26.73 | 32.42 | |
| PIV3 | Samples | 36 | 26.12 | 6.13 | 15.10 | 39.96 |
| Control | 63 | 30.81 | 1.09 | 27.60 | 33.16 | |
| PIV4 | Samples | 6 | 35.92 | 3.22 | 31.76 | 39.96 |
| Control | 22 | 33.20 | 1.39 | 29.30 | 33.99 | |
| PIV1 | Samples | 5 | 30.39 | 8.69 | 16.53 | 36.76 |
| Control | 63 | 29.93 | 0.95 | 26.23 | 32.03 | |
| PIV2 | Samples | 0 | 0.00 | 0.00 | 0.00 | 0.00 |
| Control | 63 | 30.70 | 0.88 | 26.95 | 32.89 | |
| HCoV-HKU1 | Samples | 10 | 25.82 | 5.02 | 20.66 | 37.71 |
| Control | 22 | 32.43 | 1.79 | 26.77 | 36.88 | |
| HCoV-229E | Samples | 7 | 30.41 | 3.60 | 25.98 | 35.86 |
| Control | 22 | 35.05 | 2.38 | 27.21 | 37.94 | |
| HCoV-OC43 | Samples | 7 | 25.16 | 4.64 | 17.51 | 30.88 |
| Control | 22 | 33.31 | 1.64 | 28.21 | 36.90 | |
| HCoV-NL63 | Samples | 4 | 29.69 | 4.35 | 25.67 | 34.88 |
| Control | 22 | 34.65 | 1.55 | 29.92 | 37.95 | |
| HBoV | Samples | 14 | 29.58 | 10.74 | 11.79 | 40.00 |
| Control | ||||||
| Resp/ E | Samples | 9 | 28.46 | 2.78 | 24.82 | 32.32 |
| Control | ||||||
| RNaseP | Samples | 512 | 25.29 | 3.32 | 19.66 | 37.65 |
| Control | 22 | 30.89 | 1.24 | 28.78 | 34.17 | |
Results
Of the 542 patients selected from Jan - Dec 2009 for testing for other respiratory viruses, 307 (56.7%) were males and their mean age was 3.2 years (range 0.0 to 77 years). For this analysis, patients were grouped by age as follows: 20 (3.7%) were newborns; 354 (65.3%) were infants 28 days to 1 year old; 106 (19.6%) were preschool children 2 to 6 years old; 27 (5%) were school-age children; 10 (1.8%) were school-age adolescents; and 25 (4.6%) were adults ≥ 19 years old.
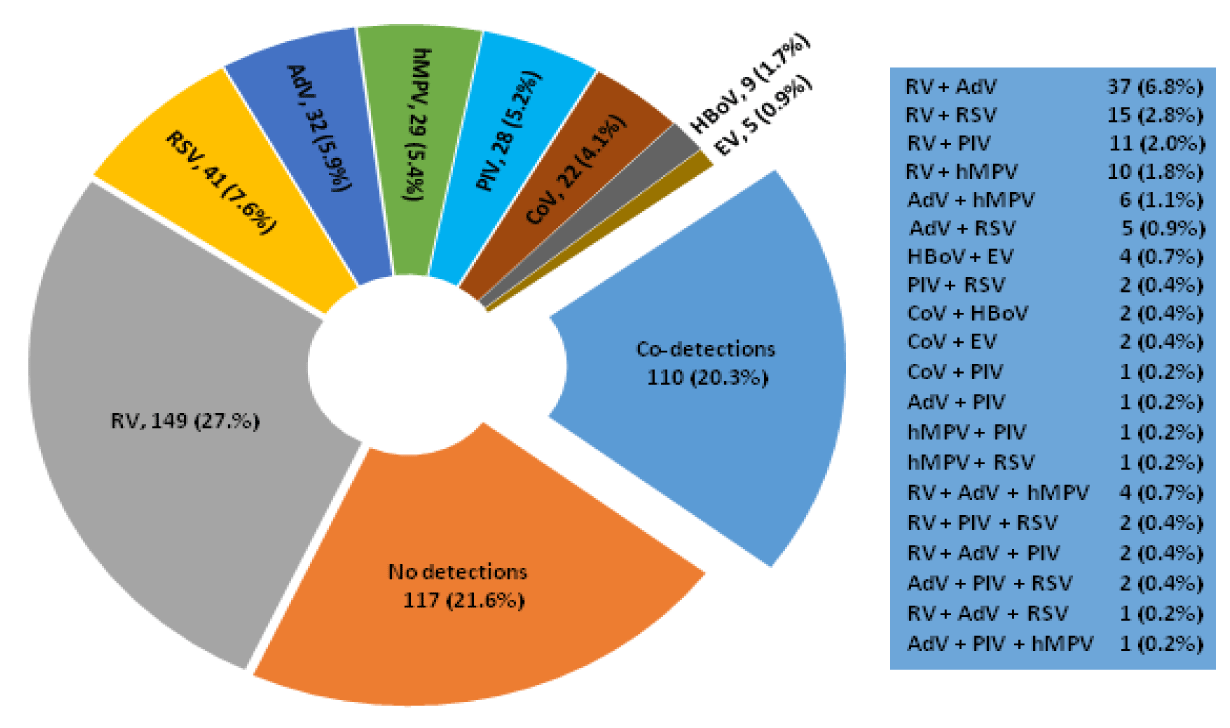
Overall, 425 of the 542 (78.4%) patients were real-time RT-PCR (RT-qPCR) positive for one or more other respiratory viruses. These included single detections of RV (149, 27.5%); RSV (41, 7.6%); AdV (32, 9.0%); HMPV (29, 5.4%); PIV (28, 5.2%); HCoV (22, 4.1%); HBoV (9, 1.7%); and EV (5, 0.9%). Patients with co-detections (110, 20.3%) of two (98, 18%) and three viruses (12, 2%) were also identified in figure 2. RV was the most common virus detected overall (27.5% of all detections) followed by RSV (7.6 % of all detections).
Of the 542 specimens negative for influenza by real-time RT-PCR( RT-qPCR) and a panel of other respiratory viruses by IIF, 219 were collected in the pre-pandemic period from January - April 2009. Of these, 114 (52.1%) were positive by RT-qPCR for respiratory viruses other than influenza. These included single detections of RV (42, 36.8%); RSV (18, 15.8%); AdV (8, 7.0%); HCoV-NL63 (3, 2.6%); EV (2, 1.75%); HMPV (1, 0.87%); PIV3 (1, 0.87%); HCoV-OC43 (1, 0.87%); HBoV (1, 0.87%); and PIV4 (1, 0.87%). HCoV-HKU1 and -229E were not identified during the pre-pandemic period. We also observed co-detections with two viruses (17, 14.91%) and three viruses (1, 0.87%). It is noteworthy that non-influenza respiratory viruses had been circulating before the emergence of the influenza pandemic strain. The Ct values of positive samples are shown in table 1.
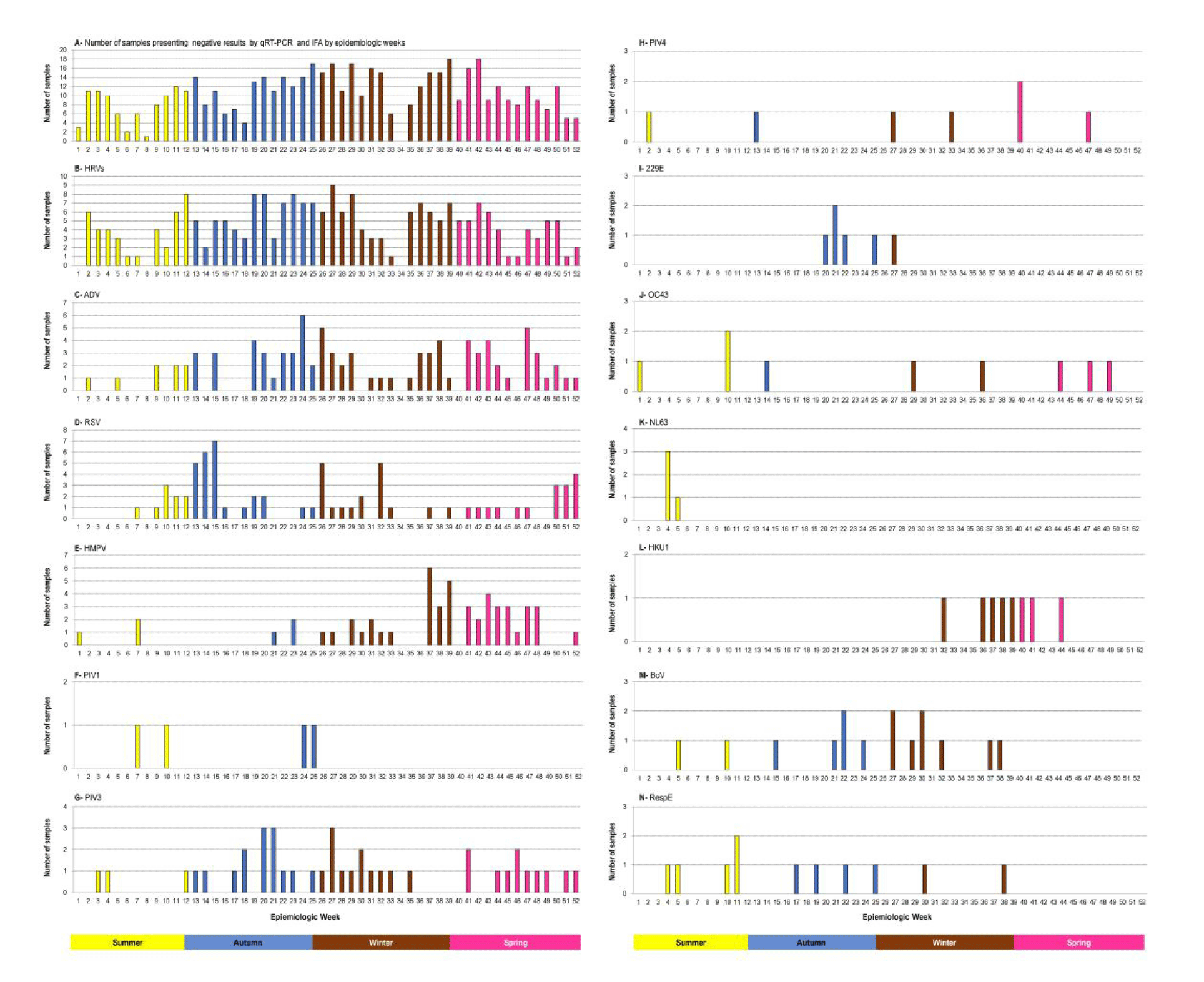
RVs were the most common viruses detected in both pre-and-pandemic periods. Molecular methodologies such as real-time RT - PCR (RT-qPCR) recovered the following non-influenza viruses from January to April 2009: HMPV, PIV4, HBoV, EV, and coronaviruses OC43, NL63 was detected only in week 5, corresponding to the summer season. Regarding coronavirus 229E, its circulation was identified from weeks 20 to 22 besides weeks 25 and 27 (autumn-winter) whereas HKU1 was detected from week 32, following weeks 36 to 41 and 44 (spring); besides those identified by IIF differential diagnosis. Due to the high specificity of the real-time RT-PCR (RT-qPCR) , RSV, AdV, PIV1, PIV2, and PIV3 viruses were also recovered from the IIF-negative samples selected for this study. The trends of seasonality of influenza viruses and other respiratory viruses, using IIF, during the pre- and pandemic periods, in 2009 are summarized in figure 4, showing the circulation of RSV, AdV, PIV1, PV3 during the pandemic period. On the other hand, PIV2 circulation was detected only in the 2009 pre-pandemic period.
A retrospective study in 2009 using real-time RT-PCR (RT-qPCR), revealed the concomitant circulation of RVs, ADV, RSV, HMPV, PIV1, PIV3, PIV4, coronaviruses 229E, OC43, and HKU1, during the pandemic period. Coronavirus NL63 circulation was observed during the pre-pandemic period as shown in figure 3. With the exception of RV which predominated in all four seasons, the following trends in the RSV and HMPV seasonality were observed: RSV peaking during autumn - winter, and HMPV peaking during spring; AdV was detected all year round as shown by IIF, although a retrospective study using real time RT - PCR (RT-qPCR) revealed an increasing activity during the winter and spring. PIV3 activity was predominant during the pandemic period in autumn–winter. The remarkable prevalence of HMPV and the low activity of PIV3 and RSV during spring provide insights concerning the seasonality of these viruses during the pandemic period (Figure 3). Apart from the activity of coronaviruses 229E and HKU1 during winter and spring, respectively, a low activity of NL3 was observed during summer, while a low activity of OC43 was noted in all four seasons (Figure 3). Although HBoV also presented low activity in autumn and early spring, its detection was concentrated during the winter period; similarly, a low detection of Resp/E was observed from autumn to early spring. The high activity of RVs in all four seasons is clearly demonstrated; these data clarify the high co-detection between RVs and other respiratory viruses as shown in figure 2.
Discussion
The results of this retrospective study revealed the high prevalence of non-influenza respiratory viruses identified in patients presenting with influenza-like illness during the influenza A (H1N1) pdm09 pandemic period. Of these, RVs were the most prevalent followed by RSV, AdV, HMPV, PIV3, PIV1, HBoV, OC43, PIV4, HKU1, Resp/E, 229E, NL63. Historically, RVs were not considered a pathogen responsible for severe respiratory diseases, although this scenario has changed after the molecular diagnosis [20-23]. According to literature data, 20 to 25% of suspected cases of influenza virus infection were diagnosed as RVs [24,25] In fact, high rates of HRV responsible for ILI outcomes, during the pandemic period, have already been described [30-36]. Tregoning JS and Schwaew J [31] have reported viral co-infections around 20%. In this retrospective study, from 542 respiratory specimens under investigation, a total of 425 tested positive as follows: 315(58%) of mono-infection, 98 (18%) due to double infection, and 12 (2%) as a result of triple infection as shown in figure 2.
A limitation regarding mixed infections with PIV4, HCoV (OC43, 229E, HKU1, NL63), HBoV, and EV also has been identified due to our laboratory routine procedures. Firstly, 542 selected samples presenting negative results for Flu by both methodologies IIF and real time RT - PCR (RT-qPCR) were investigated using the CDC real-time RT - PCR (RT-qPCR) protocol containing the following respiratory pathogens primers/probes: RSV, PIV(1, PIV2, PIV3), AdV, HMPV, and HRV. This protocol provided 170 negative results. In this context, only the negative samples were submitted to the protocol represented by primer/probes to identify the other respiratory pathogens included in the CDC real-time RT - PCR (RT-qPCR) as following PIV4, HCoV (OC43, 229E, HKU1, NL63) and EV. This work protocol has been limited to generating information regarding co-detection with PIV4, HCoV, HBoV, and Resp/E among the most prevalent viruses causing ILI clinical symptoms RSV, PIV1, PIV2, PIV3, AdV, HMPV, and HRV). A limitation regarding the detection of Resp/E and HBoV has also been observed due to the lack of Resp/E and HBoV positive control as shown in table 1.
The samples selected for investigation of non-influenza respiratory viruses were not tested for respiratory bacterial pathogens, a study limitation.
The improvement of molecular methodologies by real-time RT-PCR (RT-qPCR) multiplex platforms has contributed to identifying a large panel of respiratory viruses, other than influenza, and to clarifying the etiology of acute respiratory infection. The study clearly demonstrated that RV circulating, during 2009, did not follow a seasonal pattern which means its detection has occurred during all four seasons. In this context independently of the seasonality profile of AdV, RSV, PIVs, and HMPV virus’s co-detection with RSV has been observed as shown in figure 3. Otherwise. the co-detection between AdV and RSV was favorable due to the RSV seasonal profile during the autumn and between AdV and HMPV due to the HMPV seasonal profile at the end of winter and during the spring. It is interesting to observe that despite the low circulation of the HCoV, HBoV, and Resp/E co-detection has been observed among them during 2009.
This retrospective study demonstrates that with the exception of RVs and AdVs, the other respiratory viruses circulating in São Paulo, followed a seasonal pattern during the pandemic period as shown in figure 3. A limitation of this retrospective study concerns the lack of archived respiratory samples corresponding to the epidemiological week 34.
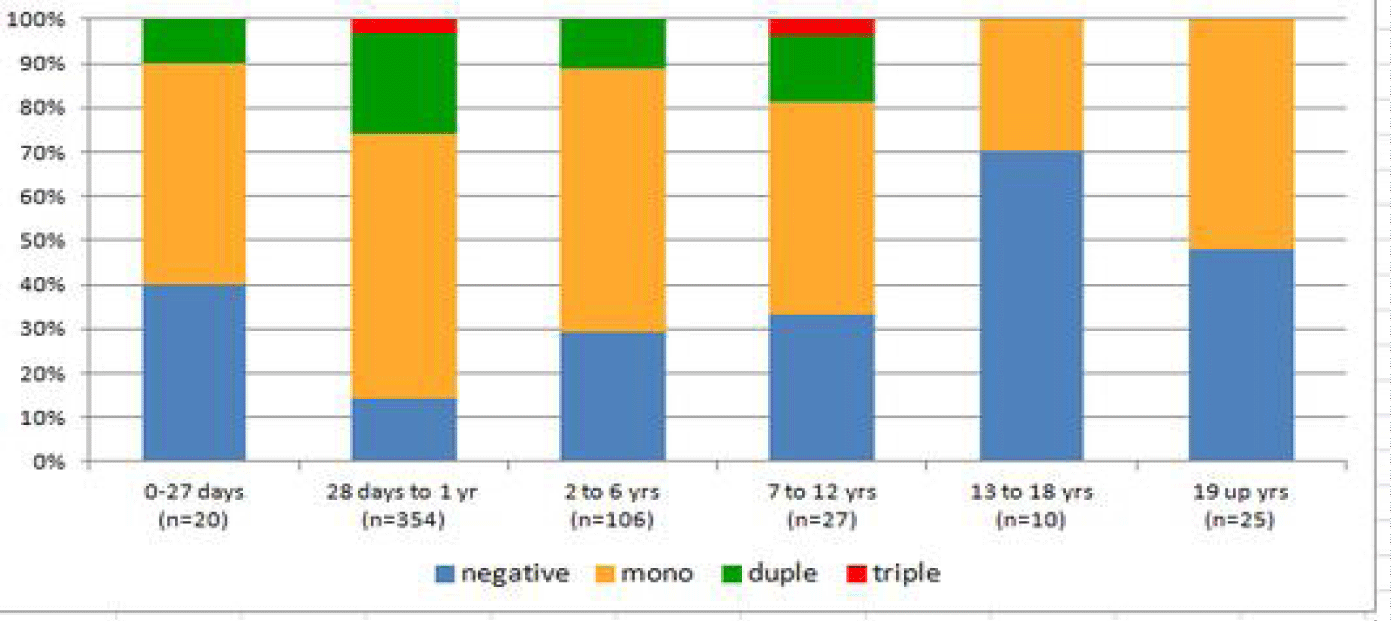
The present study shows the prevalence of viral co-detection in children under two years of age, however despite the diversity of circulation of other non-influenza respiratory viruses’ mono-infection dominates each age group as shown in figure 5.
The diversity of respiratory non-influenza viruses involved in ILI cases, during the pandemic period, certainly was revealed due to the increase in medical consultations, resulting from the panic caused by the new influenza strain.
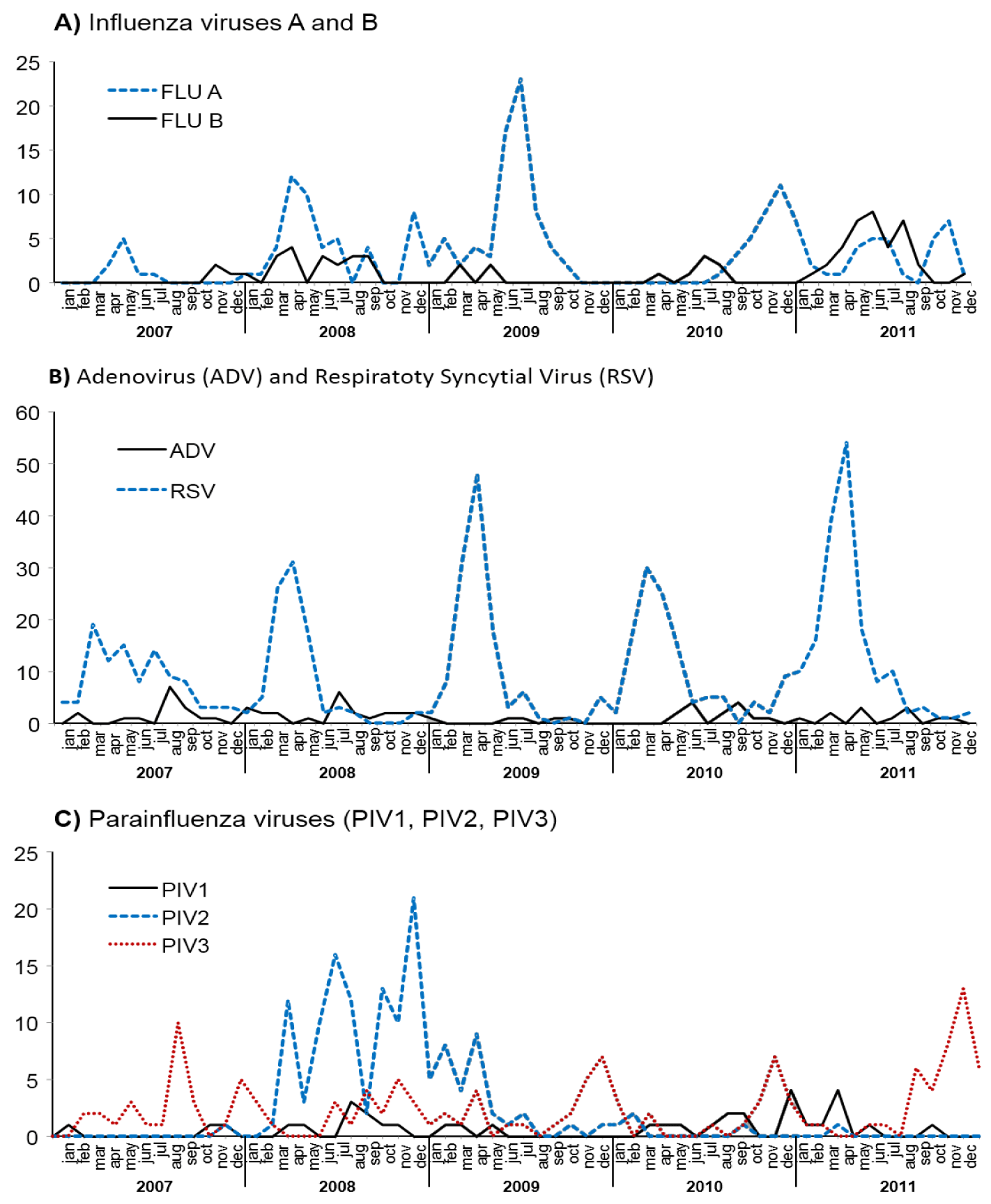
A retrospective analysis from 2007 - April 2011, based on laboratory-confirmed cases by IIF, demonstrates that RSV was the. most prevalent non-influenza virus circulating in São Paulo, with the exception of 2008 year when a remarkable PIV2 prevalence was observed. Respiratory virus differential diagnosis by using real-time RT-PCR (RT-qPCR) revealed the high prevalence of RVs circulating in 2009 in patients suspected of infection by the influenza pandemic strain. In contrast, by using the routine IIF respiratory panel RSV was the most prevalent non-influenza respiratory virus in the same period as shown in figure 4.
Considering the impact of infection by respiratory viruses in all age groups, especially among children < 5 years of age [32-34] patients with chronic diseases and adults ≤ 60 years of age, a rapid diagnosis is important to improve patient clinical management.
This retrospective study highlights how powerful real-time RT - PCR (RT-qPCR) respiratory differential diagnoses are to clarify ILI and SARI viral etiologies, during pre-and pandemic periods by recovering viruses among samples that previously tested negative by IIF and showing the real scenario of virus circulation, during a pandemic period in the city of São Paulo, as requested by the local scientific community. Our findings revealed to the Brazilian Public Health Authorities the high incidence of RVs in suspected cases of infection by the influenza pandemic strain A (H1N1) pdm09. The improvement of respiratory virus diagnosis using real-time RT - PCR (RT-qPCR) CDC protocol for other non-influenza respiratory viruses will cooperate with studies to determine the burden of the whole range of respiratory viruses involved in ILI cases, as already proposed in the global context [35]. Also, the Institute Adolfo Lutz has improved its routine respiratory virus surveillance differential diagnosis by replacing IIF with the RT --qPCR to identify RSV, AdV, PIV1, PIV2, PIV3, HMPV, and RVs based on the results from the present study. In addition, the World Health Organization (WHO) encourages epidemiological surveillance for non-influenza respiratory viruses, to identify the most prevalent viruses aiming for vaccine production. Briefly, this research will contribute to the information obtained - during the first influenza pandemic of the 21st century - from a tropical country in Latin America, the South Hemisphere, in the global scenario context.
Acknowledgment
We are indebted to Nilva Matias dos Reis, Ana Lúcia de Carvalho Avelino, Ana Maria Mota, Juliana Cristina Pereira, Norio Augusto Sasaki for the laboratory technical support. Andressa Mathias and Fabiana Cristina Pereira dos Santos for preparing the compiled results from the Excel spreadsheets. We also would like to thank the Institute Adolfo Lutz, physicians, nurses, and other health care professionals who work in the sentinel units; the Epidemiological Surveillance Centre, State Secretariat for Health São Paulo’s, Brazil; Municipal and State Secretariat for Health São Paulo, SP, Brazil; National Influenza Surveillance Network, coordinated by the Ministry of Health of Brazil (MoH). We are particularly grateful to Mr. Brett Whitaker and Dr. Teresa C.T. Peret from the Division of Viral Diseases, CDC, USA, for hands-on training in performing the RT-qPCR assays for non-influenza respiratory viruses and Dr. Dean Erdman for the critical review of this manuscript.
Competing Interests
The authors have no conflicts of interest to declare.
Funding
FAPESP – Research Project Regular, reference (2012/21922-0)
References
- WHO surveillance case definitions for ILI and SARI. WHO. 2014.
- WHO World Health Statistics. Global Health Indicators. 2013;63:95.
- Beck ET, Henrickson KJ. Molecular diagnosis of respiratory viruses. Future Microbiol. 2010 Jun;5(6):901-16. doi: 10.2217/fmb.10.48. PMID: 20521935.
- van den Hoogen BG, de Jong JC, Groen J, Kuiken T, de Groot R, Fouchier RA, Osterhaus AD. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat Med. 2001 Jun;7(6):719-24. doi: 10.1038/89098. PMID: 11385510; PMCID: PMC7095854.
- van der Hoek L, Pyrc K, Jebbink MF, Vermeulen-Oost W, Berkhout RJ, Wolthers KC, Wertheim-van Dillen PM, Kaandorp J, Spaargaren J, Berkhout B. Identification of a new human coronavirus. Nat Med. 2004 Apr;10(4):368-73. doi: 10.1038/nm1024. Epub 2004 Mar 21. PMID: 15034574; PMCID: PMC7095789.
- Peiris JS, Lai ST, Poon LL, Guan Y, Yam LY, Lim W, Nicholls J, Yee WK, Yan WW, Cheung MT, Cheng VC, Chan KH, Tsang DN, Yung RW, Ng TK, Yuen KY; SARS study group. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003 Apr 19;361(9366):1319-25. doi: 10.1016/s0140-6736(03)13077-2. PMID: 12711465; PMCID: PMC7112372.
- Ksiazek TG, Erdman D, Goldsmith CS, Zaki SR, Peret T, Emery S, Tong S, Urbani C, Comer JA, Lim W, Rollin PE, Dowell SF, Ling AE, Humphrey CD, Shieh WJ, Guarner J, Paddock CD, Rota P, Fields B, DeRisi J, Yang JY, Cox N, Hughes JM, LeDuc JW, Bellini WJ, Anderson LJ; SARS Working Group. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med. 2003 May 15;348(20):1953-66. doi: 10.1056/NEJMoa030781. Epub 2003 Apr 10. PMID: 12690092.
- Woo PC, Lau SK, Chu CM, Chan KH, Tsoi HW, Huang Y, Wong BH, Poon RW, Cai JJ, Luk WK, Poon LL, Wong SS, Guan Y, Peiris JS, Yuen KY. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J Virol. 2005 Jan;79(2):884-95. doi: 10.1128/JVI.79.2.884-895.2005. PMID: 15613317; PMCID: PMC538593.
- Allander T, Tammi MT, Eriksson M, Bjerkner A, Tiveljung-Lindell A, Andersson B. Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc Natl Acad Sci U S A. 2005 Sep 6;102(36):12891-6. doi: 10.1073/pnas.0504666102. Epub 2005 Aug 23. Erratum in: Proc Natl Acad Sci U S A. 2005 Oct 25;102(43):15712. PMID: 16118271; PMCID: PMC1200281.
- Gaynor AM, Nissen MD, Whiley DM, Mackay IM, Lambert SB, Wu G, Brennan DC, Storch GA, Sloots TP, Wang D. Identification of a novel polyomavirus from patients with acute respiratory tract infections. PLoS Pathog. 2007 May 4;3(5):e64. doi: 10.1371/journal.ppat.0030064. PMID: 17480120; PMCID: PMC1864993.
- Allander T, Andreasson K, Gupta S, Bjerkner A, Bogdanovic G, Persson MA, Dalianis T, Ramqvist T, Andersson B. Identification of a third human polyomavirus. J Virol. 2007 Apr;81(8):4130-6. doi: 10.1128/JVI.00028-07. Epub 2007 Feb 7. PMID: 17287263; PMCID: PMC1866148.
- McErlean P, Shackelton LA, Lambert SB, Nissen MD, Sloots TP, Mackay IM. Characterisation of a newly identified human rhinovirus, HRV-QPM, discovered in infants with bronchiolitis. J Clin Virol. 2007 Jun;39(2):67-75. doi: 10.1016/j.jcv.2007.03.012. Epub 2007 May 7. PMID: 17482871; PMCID: PMC7172271.
- Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012 Nov 8;367(19):1814-20. doi: 10.1056/NEJMoa1211721. Epub 2012 Oct 17. Erratum in: N Engl J Med. 2013 Jul 25;369(4):394. PMID: 23075143.
- Oliveira WK, Carmo EH, Penna GO, Kuchenbecker RS, Santos HB, Araujo WN, Malaguti R, Duncan BB, Schmidt MI. On behalf of the surveillance team for the pandemic influenza A(H1N1) 2009 in the ministry of health. Pandemic H1N1 influenza in Brazil: Analysis of the first 34,506 notified cases of influenza-like illness with severe acute respiratory infection (SARI). Euro Surveill. 2009;14(42).
- Center for Disease Control and Prevention-CDC. CDC Protocol for realtime RTPCR for influenza A (H1N1). 2009.
- Lu X, Chittaganpitch M, Olsen SJ, Mackay IA, et al. Real-Time PCR assays for detection of bocavirus in human specimens. J Clin Microbiol. 2006;44(9):3231–3235.
- Dare RK, Fry AM, Chittaganpitch M, Sawanpanyalert P, Olsen SJ, Erdman DD. Human coronavirus infections in rural Thailand: a comprehensive study using real-time reverse-transcription polymerase chain reaction assays. J Infect Dis. 2007;196(9):1321-8.
- Kodani M, Yang G, Conklin LM, et al. Application of TaqMan Low-Density Arrays for simultaneous detection of multiple respiratory pathogens. J Clin Microbiol. 2011;49(6):2175–2182.
- Weinberg GA, Schnabel KC, Erdman DD, et al. Field evaluation of TaqMan Array Card (TAC) for the simultaneous detection of multiple respiratory viruses in children with acute respiratory infection. J Clin Virol. 2013;57(3):254-60.
- Lamson D, Renwick N, Kapoor V, Liu Z, Palacios G, Ju J, Dean A, St George K, Briese T, Lipkin WI. MassTag polymerase-chain-reaction detection of respiratory pathogens, including a new rhinovirus genotype, that caused influenza-like illness in New York State during 2004-2005. J Infect Dis. 2006 Nov 15;194(10):1398-402. doi: 10.1086/508551. Epub 2006 Oct 6. PMID: 17054069; PMCID: PMC7110122.
- Arruda E, Pitkäranta A, Witek TJ Jr, Doyle CA, Hayden FG. Frequency and natural history of rhinovirus infections in adults during autumn. J Clin Microbiol. 1997 Nov;35(11):2864-8. doi: 10.1128/jcm.35.11.2864-2868.1997. PMID: 9350748; PMCID: PMC230076.
- Bellei N, Carraro E, Perosa AH, Benfica D, Granato CF. Influenza and rhinovirus infections among health-care workers. Respirology. 2007 Jan;12(1):100-3. doi: 10.1111/j.1440-1843.2006.00949.x. PMID: 17207033; PMCID: PMC7192231.
- Watanabe A, Carraro E, Kamikawa J, Leal E, Granato C, Bellei N. Rhinovirus species and their clinical presentation among different risk groups of non-hospitalized patients. J Med Virol. 2010 Dec;82(12):2110-5. doi: 10.1002/jmv.21914. PMID: 20981801.
- Nicholson KG, Kent J, Hammersley V, Cancio E. Acute viral infections of upper respiratory tract in elderly people living in the community: comparative, prospective, population based study of disease burden. BMJ. 1997 Oct 25;315(7115):1060-4. doi: 10.1136/bmj.315.7115.1060. PMID: 9366736; PMCID: PMC2127683.
- Boivin G, Osterhaus AD, Gaudreau A, Jackson HC, Groen J, Ward P. Role of picornaviruses in flu-like illnesses of adults enrolled in an oseltamivir treatment study who had no evidence of influenza virus infection. J Clin Microbiol. 2002 Feb;40(2):330-4. doi: 10.1128/JCM.40.2.330-334.2002. PMID: 11825938; PMCID: PMC153349.
- Gunson RN, Carman WF. During the summer 2009 outbreak of "swine flu" in Scotland what respiratory pathogens were diagnosed as H1N1/2009? BMC Infect Dis. 2011 Jul 13;11:192. doi: 10.1186/1471-2334-11-192. PMID: 21752259; PMCID: PMC3146830.
- Casalegno JS, Ottmann M, Duchamp MB, Escuret V, Billaud G, Frobert E, Morfin F, Lina B. Rhinoviruses delayed the circulation of the pandemic influenza A (H1N1) 2009 virus in France. Clin Microbiol Infect. 2010 Apr;16(4):326-9. doi: 10.1111/j.1469-0691.2010.03167.x. Epub 2010 Jan 28. PMID: 20121829.
- Nisii C, Meschi S, Selleri M, Bordi L, Castilletti C, Valli MB, Lalle E, Lauria FN, Piselli P, Lanini S, Ippolito G, Di Caro A, Capobianchi MR. Frequency of detection of upper respiratory tract viruses in patients tested for pandemic H1N1/09 viral infection. J Clin Microbiol. 2010 Sep;48(9):3383-5. doi: 10.1128/JCM.01179-10. Epub 2010 Jun 30. PMID: 20592147; PMCID: PMC2937695.
- Ratnamohan VM, Taylor J, Zeng F, McPhie K, Blyth CC, Adamson S, Kok J, Dwyer DE. Pandemic clinical case definitions are non-specific: multiple respiratory viruses circulating in the early phases of the 2009 influenza pandemic in New South Wales, Australia. Virol J. 2014 Jun 18;11:113. doi: 10.1186/1743-422X-11-113. PMID: 24942807; PMCID: PMC4076060.
- Schnepf N, Resche – Rigon M, Chaillon A, Scemla A, Gras G, Semoun O, Taboulet P, Molina JM, Simon F, Goudeau A, LeGoff J. High burden of non-influenza viruses in influenza – like illness in the early of H1N1v Epidemic in France. Plos one. 2011;6(8):e23514.
- Tregoning JS, Schwarze J. Respiratory viral infections in infants: causes, clinical symptoms, virology, and immunology. Clin Microbiol Rev. 2010 Jan;23(1):74-98. doi: 10.1128/CMR.00032-09. PMID: 20065326; PMCID: PMC2806659.
- Nascimento-Carvalho CM, Ribeiro CT, Cardoso MR, Barral A, Araújo-Neto CA, Oliveira JR, Sobral LS, Viriato D, Souza AL, Saukkoriipi A, Paldanius M, Vainionpää R, Leinonen M, Ruuskanen O. The role of respiratory viral infections among children hospitalized for community-acquired pneumonia in a developing country. Pediatr Infect Dis J. 2008 Oct;27(10):939-41. doi: 10.1097/INF.0b013e3181723751. PMID: 18756190.
- Luksic I, Kearns PK, Scott F, Rudan I, Campbell H, Nair H. Viral etiology of hospitalized acute lower respiratory infections in children under 5 years of age – a systematic review and meta – analysis. Croatian Medical Journal. 2013;54(2):122-134.
- Broor S, Dawood FS, Pandey BG, Saha S, Gupta V, Krishnan A, Rai S, Singh P, Erdman D, Lal RB. Rates of respiratory virus-associated hospitalization in children aged <5 years in rural northern India. J Infect. 2014 Mar;68(3):281-9. doi: 10.1016/j.jinf.2013.11.005. Epub 2013 Nov 21. PMID: 24269675; PMCID: PMC7112698.
- Tang JW, Lam TT, Zaraket H, Lipkin WI, Drews SJ, Hatchette TF, Heraud JM, Koopmans MP; INSPIRE investigators. Global epidemiology of non-influenza RNA respiratory viruses: data gaps and a growing need for surveillance. Lancet Infect Dis. 2017 Oct;17(10):e320-e326. doi: 10.1016/S1473-3099(17)30238-4. Epub 2017 Apr 28. Erratum in: Lancet Infect Dis. 2017 Jul;17(7):689. PMID: 28457597; PMCID: PMC7164797.
Content Alerts
SignUp to our
Content alerts.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.