Medicine Group. 2023 December 14;4(12):1664-1666. doi: 10.37871/jbres1848.
Long-term Pseudo-Hypertriglyceridemia due to Novel GK Splice Variant c.894+2T>C
Enrique Nogueira1,2*, Betariz del Olmo2, Concepción Lobo2, Cecilia González2, Patricia Llovet3 and Cristina Martínez3
2Molecular Diagnostics Laboratory, Eurofins Megalab, Madrid, Spain
3Clinical Analysis Laboratory, Eurofins Megalab, Madrid, Spain
Introduction
We here report on a case of a 46-year-old male under evaluation for persistent long-term (for more than 20 years) Hypertriglyceridemia (HTG) submitted to us for a genomic study based in an exome analysis of a mixed dyslipidemia, with Hypercholesterolemia (HC) and HTG (in multiple serum determinations, in recent years on Abbott Alinity Chemistry Analyzers), with unequal response to treatment (with statins for HC, and fibrates for HTG), with a progressive decrease in cholesterol values (now at almost optimal values of LDL cholesterol, around 125 mg/dL) while with no effect on TGs measurements (commonly around 600 mg/dL).
A clinical exome analysis, with reagents and equipments from Agilent (for SureSelect Human All Exon V8; https://www.agilent.com) and Illumina (https://emea.illumina.com), and software from Agilent, Illumina and Genoox (https://franklin.genoox.com), was undertaken searching for gene mutations of different types (sequence variants as well as exon deletions and duplications) in the entire gene coding sequences (including short intronic sequences closed to coding exons). Such a complete analysis did not lead to identification of relevant findings in genes associated to Familial HC (FHC), including three genes responsible for a majority (~90%) of cases, LDLR (60%-80%), APOB (1%-5%) and PCSK9 (up to 3%). Neither in genes occasionally implicated in FHC, most notably LDLRAP1, CYP27A1, APOE, ABCG5, ABCG8, LPL, USF1, SORT1, STAP1, LIPA and PNPLA5 [1-3]. Accordingly, an involvement of as yet unidentified genes cannot be ruled out, nor a case of polygenic origin, which after some speculations would be responsible for up to ~40% of FHC cases without an identified one-gene Mendelian linkage.
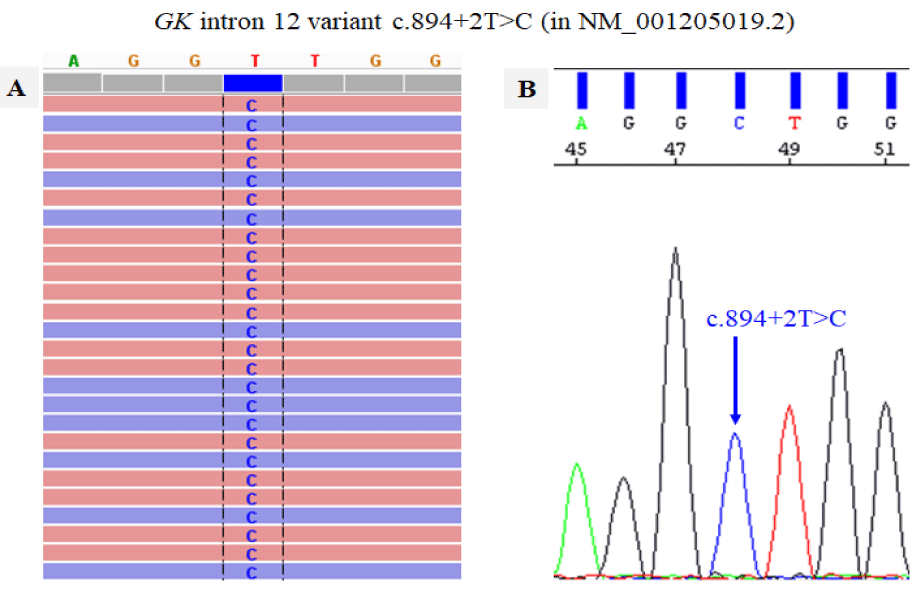
Further, a survey of genes implicated in Familial HTG (FHTG) neither yielded relevant findings. FHTG is a dyslipidemia with proposed monogenic linkage in severe cases to the genes LPL, APOC2, APOA5, LMF1, GPIHBP1 and GPD1. And with a multigene (polygenic) association in mild-moderate cases to LPL, APOC2, GCKR, APOB, LMF1, GPIHBP1, CREBH, APOE and less relevant variants of other genes [4]. However, a consideration of many other candidate genes, associated to HTG, as proposed by HPO database (https://hpo.jax.org/app/browse/term/HP:0002155), led to identification of variant c.894+2T>C of GK gene (Figure 1) considered relevant.
GK (Glycerol Kinase) is a gene of locus Xp21.2 that catalyzes the phosphorylation of glycerol to glycerol-3-phosphate, necessary for triglyceride production. It is implicated by X-Linked Recessive inheritance (XLR) in the Glycerol Kinase Deficiency (GKD), a disorder characterized by elevated plasma and urine glycerol levels and variable neurometabolic manifestations. It can occur as a contiguous gene deletion syndrome (from Xp21.2-Xp21.3) with congenital adrenal hypoplasia and Duchenne muscular dystrophy, in 'complex' forms, the severity depending on the size of the deletion (number of missing genes). In cases of exclusive GK involvement is detected only as hyperglycerolemia, as observed in our patient, with recent results of very high glycerol blood content (3,922 µmol/L; measured with a spectrophotometric assay, with reference range of 20-250 µmol/L). It becomes now clear that the high glycerol content has been the reason for the repeatedly erroneous determinations of TGs in serum, as well as in urine (37,742 mg/dL) that was tested as a proof of false determinations because TGs are never found in urine [5]. Besides, and according to a NMR spectroscopy assay (Liposcale), our patient has normal TGs values in serum, of 84 mg/dL (<150 mg/dL).
The identified GK variant, c.894+2T>C, of intron 12 (in reference sequence NM_001205019.2), is found in hemizygosis (in all the single sequences of the patient's X chromosome, as shown in Fig. 1). Although we are not aware of its occurrence before, neither in patients nor in healthy individuals, it is considered relevant, by altered mRNA splicing, as similar mutations in these locations, so that according to ACMG (American College of Medical Genetics and Genomics) criteria it could be considered as a likely pathogenic mutation, as proposed by knowledge bases Franklin (https://franklin.genoox.com/clinical-db/variant/snp/chrX-30725717-T-C) and VarSome (https://varsome.com/variant/hg19/NM_001205019:c.894+2T>C).
Thus, and in conclusion, the valuable genomic analysis of HTG based on NGS has facilitated the finding of novel variant c.894+2T>C of GK gene, responsible for the pseudo-HTG of our patient, a rare case of hyperglycerolemia due to GKD that –as it has been repeatedly described– in adulthood is often asymptomatic [6-8] and does not require pharmacological treatment, therefore avoiding the use of fibrates, although a diet at more frequent intervals and rich in complex carbohydrates might be advisable [8]. The genomic study has on the other hand not allowed us to establish a gene linkage for the HC, apparently well managed with statins which nevertheless might be related to the slight transaminase elevations noted in the patient.
References
- Ison HE, Clarke SL, Knowles JW. Familial Hypercholesterolemia. 2014 Jan 2 [updated 2022 Jul 7]. In: Adam MP, Feldman J, Mirzaa GM, Pagon RA, Wallace SE, Bean LJH, Gripp KW, Amemiya A, editors. GeneReviews® [Internet]. Seattle (WA): University of Washington, Seattle; 1993–2023. PMID: 24404629.
- De Castro-Orós I, Pocoví M, Civeira F. The genetic basis of familial hypercholesterolemia: inheritance, linkage, and mutations. Appl Clin Genet. 2010 Aug 5;3:53-64. doi: 10.2147/tacg.s8285. PMID: 23776352; PMCID: PMC3681164.
- Sharifi M, Futema M, Nair D, Humphries SE. Genetic Architecture of Familial Hypercholesterolaemia. Curr Cardiol Rep. 2017 May;19(5):44. doi: 10.1007/s11886-017-0848-8. PMID: 28405938; PMCID: PMC5389990.
- Hegele RA, Ginsberg HN, Chapman MJ, Nordestgaard BG, Kuivenhoven JA, Averna M, Borén J, Bruckert E, Catapano AL, Descamps OS, Hovingh GK, Humphries SE, Kovanen PT, Masana L, Pajukanta P, Parhofer KG, Raal FJ, Ray KK, Santos RD, Stalenhoef AF, Stroes E, Taskinen MR, Tybjærg-Hansen A, Watts GF, Wiklund O; European Atherosclerosis Society Consensus Panel. The polygenic nature of hypertriglyceridaemia: implications for definition, diagnosis, and management. Lancet Diabetes Endocrinol. 2014 Aug;2(8):655-66. doi: 10.1016/S2213-8587(13)70191-8. Epub 2013 Dec 23. PMID: 24731657; PMCID: PMC4201123.
- Fernández-Pardo J, Quílez-Fernández JL, Pedro-Botet J. Screening for hyperglycerolemia by triglyceride assay in urine. J Clin Lipidol. 2019 Sep-Oct;13(5):854-855. doi: 10.1016/j.jacl.2019.09.006. Epub 2019 Sep 14. PMID: 31604684.
- Arrobas-Velilla T, Mondéjar-García R, Gómez-Gerique JA, Cañizares Díaz I, Cruz Mengibar MC, Orive de Diego A, Fabiani-Romero F. Pseudo-hypertriglyceridaemia or hyperglycerolemia? Clin Investig Arterioscler. 2013 Jul-Aug;25(3):123-6. doi: 10.1016/j.arteri.2013.05.005. Epub 2013 Jul 19. PMID: 23877006.
- Pant V, Pyakurel D, Gautam K, Pradhan S. Pseudo-hypertriglyceridaemia in glycerol kinase deficiency misdiagnosed and treated as true hypertriglyceridaemia. BMJ Case Rep. 2022 Mar 15;15(3):e248251. doi: 10.1136/bcr-2021-248251. PMID: 35292548; PMCID: PMC8928288.
- Arrieta F, Ojeda S, Rueda Á, Stanescu S, Belanger-Quintana A, Martínez-Pardo M. Déficit de glicerol kinasa en el adulto: hipertrigliceridemia resistente a tratamiento dietético y farmacológico [Glycerol kinase deficiency in adult patient: hypertriglyceridemia resistance to diet and pharmacological treatment]. Nutr Hosp. 2018 Aug 2;35(4):993-995. Spanish. doi: 10.20960/nh.1921. PMID: 30070892.
Content Alerts
SignUp to our
Content alerts.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.