Medicine Group. 2024 February 07;5(2):117-134. doi: 10.37871/jbres1878.
Spin Practices and Low Reporting Quality in Studies on Prediction Model of Diagnosis Based on TCM Information: A Systematic Review and Evidence Mapping
Jieyun Li1, Jiekee Lim1, Xinang1, Mi Zhou1 and Zhaoxia Xu1,2*
2Shanghai key laboratory of health identification and evaluation, Shanghai, China
- Traditional Chinese medicine
- Prediction model
- Reporting standards
- Methodological quality
- Spin practice
Abstract
Background: Despite the abundance of published studies on prediction models for diagnosing Traditional Chinese Medicine (TCM), there remains a lack of comprehensive assessment regarding reporting and methodological quality, as well as an absence of examination into the objectivity of linguistic aspects within these studies.
Methods: The PubMed, Cochrane Library, Web of Science, CNKI, VIP, and WANFANG databases were systematically searched from inception to October 30th, 2023, to identify studies developing and/or validating diagnostic and prognostic TCM prediction models using supervised machine learning. PROBAST and TRIPOD were employed to assess the reporting and methodological quality of identified studies. A previous article about spin in prognostic factor studies already identified several practices, which we modified for our data extraction the present study was registered on PROSPERO with the registration number CRD42023450907.
Results: 35 and 19 eligible studies published in Chinese and English were identified respectively from 1746 records. The clinical diseases with the most publications were diabetes (n = 7, 14.8%), coronary heart disease (n = 6, 11.1%), and lung cancer (n = 5, 9.26%). Primary analysis and sensitivity analysis confirmed that the reporting and methodological quality of included studies were correlated (rs = 0.504, p < 0.001). The quality of the CM prediction model requires improvement by including a structured title, participants and predictor’s selection, statistical analysis methods, model performance and interpretation. Two studies (4.55%) recommended the model to be used in daily practice lacked any external validation of the developed models. Six studies (13.63%) made recommendations for clinical use in their main text without any external validation. Reporting guidelines were only cited in one study (1.85%).
Conclusion: The available evidence indicated TCM information can provide predict information for different diseases, but the scientific quality of published studies needs to be improved.
Introduction
The clinical method of four diagnoses, involving inspection, auscultation and olfaction, inquiry, and palpation, is emphasized in Traditional Chinese Medicine (TCM) [1]. Collecting TCM clinical indices holds important guiding significance for subsequent diagnosis and treatment conducted by TCM doctors. In general, a variety of information is required to arrive at an accurate clinical diagnosis including the patient’s medical history, physical examination, symptoms, signs, etc. With the rapid development of information technology and the popularization of big data, the modernization of diagnostic approaches has become integral to TCM [2]. In order to improve the accuracy and reliability of TCM diagnosis, so as to decrease some challenges, such as high subjectivity, strong empirical dependence, and inconsistent diagnostic outcomes, researchers have delved into harnessing machine learning and data mining technology to establish TCM prediction models that can assist in the diagnosis of various diseases in clinical practice. For instance, Wang, et al. [3], stated that the machine learning classification model based on tongue and pulse data exhibits glorious performance and can adequately predict PCOS risk; Shi, et al. [4] investigated the tongue image features among patients with lung cancer and constructed a lung cancer risk warning model using machine learning methods.
Since the publication of the Transparency Report on Multivariate Prediction Models for Individual Prognosis or Diagnosis (TRIPOD) statement [5] and the Predictive model Risk of Bias Assessment Tool (PROBAST) [6], the publication of machine learning clinical prediction methods rapidly increases. Nonetheless, studies using machine learning techniques often face questions about their actual effectiveness within the clinical workflow. In recent years, a large number of prediction models have been published, but often with inadequate reporting and methodological quality. For example, Li et al. [7], used PROBAST to assess the methodological quality of published systematic reviews of predictive models for motor function in patients 3-6 months after a stroke. Their assessment highlighted limitations, particularly the failure to report key information during predictive model development and validation. Despite the seeming objectivity and straightforwardness of prediction models based on readily available clinical information, their potential to provide risk estimates for diverse outcomes is hindered by a lack of comprehensive reporting. This limitation restricts the utility of study findings, impeding subsequent validation endeavors, evidence synthesis, or in daily practice, leading to research waste.
In addition, a methodology study focusing on predictive models has highlighted that the misuse of lingual, whether intentional or unintentional, affects the interpretation of findings and has been described as “rotation” [8]. Spin is sometimes referred to as “scientific hype,” a situation where scientific findings are inappropriately exaggerated and widely understood as a biased presentation [9]. In the scientific literature, spin refers to the specific reporting practice of distorting the interpretation of a result, thereby misleading the reader into perceiving the outcome more favorably. Evidence suggests that “spin” or over-exaggeration of scientific findings is prevalent across various study types, including randomized therapeutic interventions, observational studies, biomarker analyses, diagnostic test accuracy studies, prognostic factor investigations, and systematic reviews. This phenomenon significantly influences reader interpretation and decision-making [10].
There is no comprehensive summary of the evidence reported in TCM diagnostic prediction models, along with an evaluation of the scientific quality of published studies. Thus, we present the outcomes by a systematic review to summarize both the reporting and methodological quality of TCM diagnosis prediction model studies, in addition to examining spin practices. Moreover, we provide an evidence gap map based on Area Under the ROC Curve (AUC) results for each included study, addressing the knowledge gaps identified in this realm.
Methods
For the reporting of this study, we adhered to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) 2020 statement [11]. To increase the transparency of the research process and avoid selective reporting of results, we registered the systematic review on the International prospective register of systematic review (PROSPERO, https://www.crd.york.ac.uk/PROSPERO/), with registration number CRD42023450907. Ethical approval was waived as the study design is an overview of published systematic reviews with meta-analyses and, as such, did not involve additional human participation.
Literature search
A comprehensive and systematic search was performed to identify primary studies on machine learning-based prediction models of TCM diagnostic. Six large electronic databases, PubMed (https://pubmed.ncbi.nlm.nih.gov/), Web of Science (https://www.webofscience.com/), the Cochrane Library (https://www.cochranelibrary.com/), VIP (http://www.cqvip.com/), WANFANG (https://www.wanfangdata.com.cn/), and CNKI (https://www.cnki.net/), were searched from inception to October 30th, 2023, with Medical Subject Heading and keyword terms that included “Machine Learning”, “Deep learning”, “ Predict*”, “ Validat*”, “Medicine, Chinese Traditional”, “Complem entary Therapies”, “diagnostic”, and “ Prognostic”. The reference lists of all selected systematic reviews with meta-analyses were manually searched to identify additional articles potentially meeting the inclusion criteria. The details of all search strategies used in this review are presented in Supplementary Material.
Literature selection
We included studies that met any of the following criteria: 1) described the development or validation of one or more CM multivariable prediction models using any machine learning technique aiming for individualized predictions, or 2) reported on the incremental value or model extension aiming to develop a CM indicator-based prediction model. A multivariable prediction model was defined as a model aiming to predict a health outcome by using two or more predictor variables. We excluded studies that: 1) investigated single predictor, test, or biomarker or its causality with an outcome; 2) employed machine learning to enhance the images or signals interpretation; 3) utilized predictors solely for genetic traits or molecular markers; 4). utilized predictors solely for the effectiveness of specific CM interventions. The search was restricted to human subjects, English and Chinese language articles, and those accessible through our institution's resources. As meta-analysis essentially involves the statistical synthesis of findings from individual studies [12], preclinical studies, qualitative systematic reviews, narrative reviews, protocols, duplicate records, and publications of non-target intervention were excluded. All records identified from electronic databases were imported into Endnote (Version X9, Clarivate Analytics) to assess eligibility. Firstly, two independent reviewers (JK.Lim and XA.Xiao) screened records based on their titles and abstracts, after deduplication. Subsequently, the full text of potentially eligible records was then downloaded for further scrutiny. Any disagreement during the selection process was solved through discussion or consultation with the third author (ZX. Xu).
Quality assessment
The TRIPOD Statement consists of a comprehensive checklist comprising 22 items that are deemed crucial for the comprehensive reporting of studies focused on the development or validation of multivariable prediction models. These items encompass various aspects, spanning from the title and abstract (items 1 and 2), background and objectives (item 3), methods (items 4 through 12), results (items 13 through 17), discussion (items 18 through 20), and other information (items 21 and 22). TRIPOD explicitly covers the development and validation of prediction models for both diagnosis and prognosis, spanning all medical domains and all types of predictors [5]. PROBAST, on the other hand, consists of 4 domains (participants, predictors, outcome, and analysis), containing 20 signaling questions to facilitate Risk of Bias (ROB) assessment. The tool is now widely used as a general instrument for appraising the methodological quality of primary studies that develop, validate, or update (for example, extend) multivariable prediction models for diagnosis or prognosis [6].
‘Spin practice’ is defined as any issue that could make the clinical utility of the developed or validated prediction model look more favorable than the study design and results can underpin. A previous article about spin in prognostic factor identified several practices, which we considered for use in our data extraction [13]. For a detailed description of extracted items.
Two investigators (JK.Lim and XA.Xiao) independently evaluated the methodological and reporting quality of the studies included in the overview. Specifically, "Yes" (Y), "Partial Yes" (PY), and "No" (N) were used to answer the questions related to items in the TRIPOD. "Low ROB", "Unclear ROB", and "High ROB" were used to answer the questions related to items in PROBAST tools. The results of the assessment were cross-checked, and consensus was reached among the investigators. Any disagreement between them arising from discussions was resolved by consultation with the third author (ZX. Xu).
Data extraction
The general information encompassed in the extracted data including the title of publication, first author’s name, year of publication, writing language, journal title, study type (diagnosis vs prognosis), total sample size, the aim of the study (development only or development with external validation), clinical diseases, CM diagnosis methods, the mention of CM syndrome differentiation, the use of machine learning, and the reference to reporting guideline. The extraction form was pilot tested on five articles and was subsequently implemented using Excel 2019 (Microsoft Corporation, WA, USA). Two investigators (JK.Lim and XA.Xiao) independently extracted key outcome information and the study characteristics from each document using a predesigned Excel spreadsheet. Any disagreement was resolved through discussion or by consultation with the third author (ZX.Xu).
Statistical analysis
Data relating to prediction outcomes of various machine learning based on CM indicators for clinical diseases in the included studies were descriptively summarized. The compliance rate of TRIPOD statement items was calculated for each study and reported the numbers and their percentages of "Y", "PY", or "N" responses. Meanwhile, the numbers and percentages of "Low ROB", "unclear", or "High ROB" responses were calculated for PROBAST items. Several evidence mapping methods were used to visualize the results of quality assessment [14,15]. A radar plot was used to present the assessment of reporting quality, while a bar chart was used to display the results of methodological quality. Meanwhile, a bubble plot was used to visualize the results of the overall methodological quality of each included study. Each bubble represented a primary study, the size of each bubble proportional to sample size (ie., total number of participants), and the color of the bubble represented the disease specialty classified based on ICD-11 (International classification of diseases-11) [16]. The X-axis reflected the best AUC score for internal validation of the prediction model study, whereas the Y-axis presented the overall methodological quality assessed by the PROBAST. The overall confidence of each study was classified as “Critically Low” (more than one critical flaw with or without non-critical weaknesses), “Low” (one critical flaw with or without non-critical weaknesses), “Moderate” (more than one non-critical weakness), or “High” (no or one non-critical weakness). Two reviewers (JY.Li and M.Zhou) with a background in evidence-based medicine, employed this tool to independently evaluate the methodological quality of the included studies, while any further disagreement was resolved through detailed discussions.
In addition, Spearman's rank (rs) test was employed to explore the correlation between reporting quality and methodological quality using the number of responses in TRIPOD and PROBAST items for each study. To assess the robustness of the primary analysis, sensitivity analysis was conducted after excluding the "PY” and “unclear” responses. The strength of correlation was rated as low (rs < 0.4), moderate (rs: 0.4~0.7), and high (rs > 0.7) [17]. Excel 2019 and Stata 16.0 (StataCorp, College Station, TX, USA) were used to analyze and visualize the data. Statistical significance was set as two-sided p < 0.05.
Results
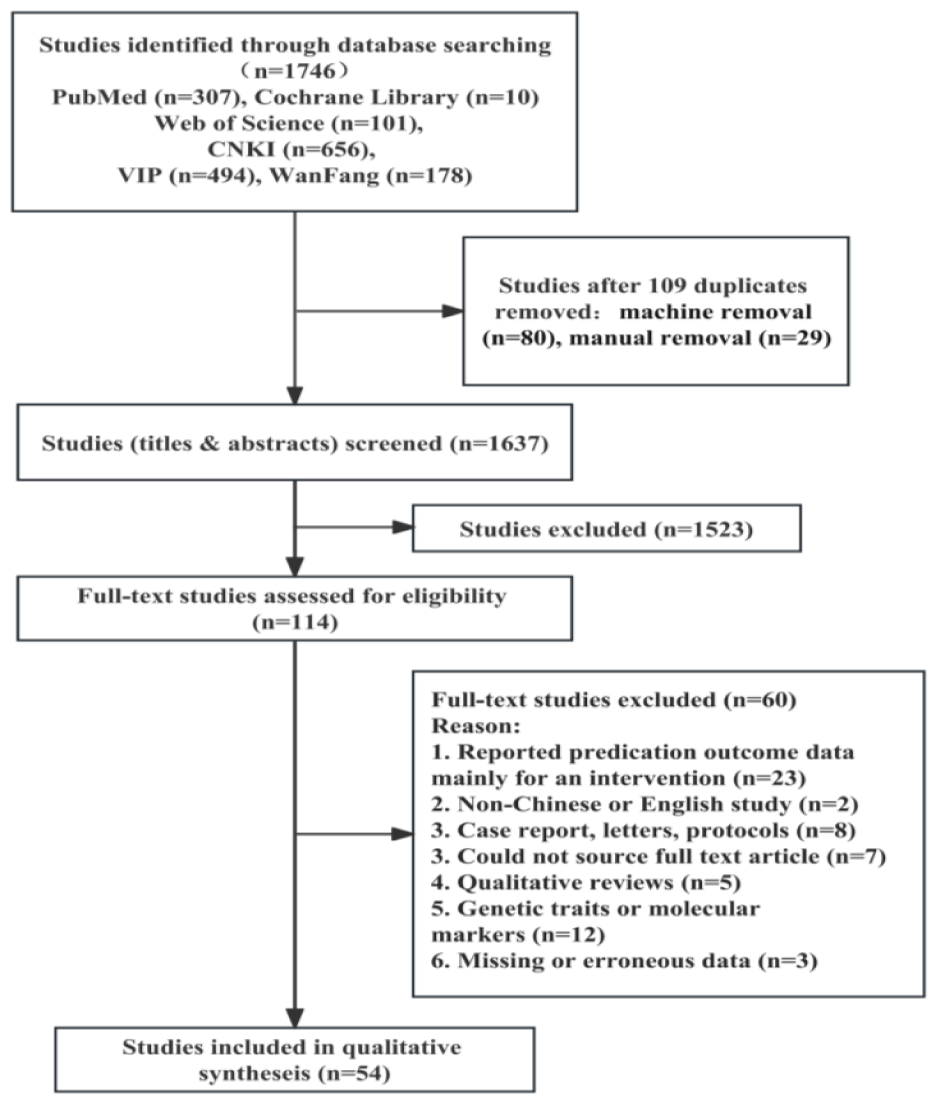
A total of 1746 records were identified from six databases (PubMed = 307, Web of Science = 101, The Cochrane Library =10, CNKI = 656, VIP = 494, WANFANG =178). 1637 records were screened after removing the 109 duplications. Through the meticulous evaluation of the titles and abstracts, a further 1523 records were excluded. Ultimately, 54 studies [3,4,18-69] focusing on machine learning-based prediction models for CM diagnostic were selected for final inclusion in the research. A flow diagram of the study selection process is presented in figure 1.
General characteristics of included studies
Table 1 shows the characteristics of the included articles. All 54 articles originated from Chinese institutions, of which 19 (35.19%) were written in English and 35 (64.81%) in Chinese. 32 studies (59.26%) were published in 16 various journals, 15 studies (27.78%) were published in master dissertations, and 7 (12.96%) for doctoral dissertations. The broad spectrum of total sample sizes spanned from 12,000 as the largest to 98 as the smallest. Three studies (5.5%) focused on prognostic models [45,53,56] and 51 (94.44%) on diagnostic models. Most studies reported the development of prediction models, including internal validation (n = 44, 81.5%), and 10 (18.5%) performed external validation. Among the clinical diseases under investigation, the most prevalent were diabetes (n = 7, 14.8%) [20,22,23,34,44,57,62], Coronary Heart Disease (CHD) (n = 6, 11.1%) [18,31,37,43,50,67], and lung cancer (n = 5, 9.26%) [4,29,36,39,53]. 31 studies (57.41%) used conventional CM four-diagnosis information, including inquiry scale, tongue diagnosis and pulse diagnosis, which binary data were used for prediction. Moreover, 9 studies (16.67%) used objectified parameters collected by tongue instrument and 12 studies (22.22%) by pulse-taking instrument. 29 studies (53.7%) predicted the outcome of different CM syndrome types. Support Vector Machine (SVM) and Random Forest (RF) were the most widely used machine learning, utilized in 28 (51.85%) and 22 (40.74%) instances, respectively. Remarkably, the citation of reporting guidelines was observed in only one study (1.86%) [61], which mentioned TRIPOD.
| Table 1: General characteristics of included articles (n = 54). | |||||||||||
| Code | Author | Language | Publication | Study type | Sample Size | Study aim | Clinical disease | CM predictive factor | CM syndrome differentiation | Machine learning | Reference to reporting guideline |
| 1 | [66] | Chinese | J of CM | Diagnosis | 1021 | Development only | Menopause | Conventional CM information | NR | RF; SVM; ANN | NR |
| 2 | [58] | Chinese | J of Zhejiang Chinese Medical University | Diagnosis | 305 | Development with external validation | Stroke | Conventional CM information | yes | LR | NR |
| 3 | [54] | Chinese | J of Hunan University of CM | Diagnosis | 300 | Development only | Stroke | Conventional CM information | yes | SVM; KNN; RF; ExtraTree; XGBoost; LightGBM | NR |
| 4 | [4] | English | Front Physiol | Diagnosis | 862 | Development only | Lung cancer | Tongue instrument parameters | NR | DT; SVM; RF; NN; NB; LR | NR |
| 5 | [3] | English | Digit Health | Diagnosis | 486 | Development only | PCOS | Pulse-taking instrument parameters; Tongue instrument parameters | NR | AdaBoost; SVM | NR |
| 6 | [18] | English | Comput Math Methods Med | Diagnosis | 238 | Development only | CHD | Laboratory test indicators | yes | ANN | NR |
| 7 | [69] | Chinese | J of Yanshan University | Diagnosis | 906 | Development only | Corporeity | Tongue instrument parameters | NR | LR; RF; XGBoost; AdaBoost | NR |
| 8 | [64] | Chinese | Intelligent computers and applications | Diagnosis | 2520 | Development only | Corporeity | Tongue instrument parameters | NR | LR; ANN; SVM; NB | NR |
| 9 | [61] | Chinese | Masteral dissertation | Diagnosis | 415 | Development only | COPD | Conventional CM information | yes | NN | TRIPOD |
| 10 | [39] | Chinese | Masteral dissertation | Diagnosis | 350 | Development only | Lung cancer | Conventional CM information | NR | CNN | NR |
| 11 | [61] | Chinese | Masteral dissertation | Diagnosis | 235 | Development with external validation | MCI | Conventional CM information | yes | LR; XGBoost | NR |
| 12 | [68] | Chinese | Masteral dissertation | Diagnosis | 845 | Development only | Eczema | Pulse-taking instrument parameters | yes | SVM; RF; ANN | NR |
| 13 | [50] | Chinese | Tianjin CM | Diagnosis | 149 | Development only | Hypertension | Pulse-taking instrument parameters;Tongue instrument parameters | NR | LR | NR |
| 14 | [67] | Chinese | Doctoral dissertation | Diagnosis | 1676 | Development with external validation | Cardiac failure | Conventional tongue information | yes | RF; DT | NR |
| 15 | [49] | Chinese | Masteral dissertation | Diagnosis | 500 | Development with external validation | Hypertension | Conventional CM information | yes | LR | NR |
| 16 | [50] | Chinese | J of CM | Diagnosis | 4723 | Development with external validation | Diabetes | Conventional tongue information | NR | NB | NR |
| 17 | [42] | Chinese | Masteral dissertation | Diagnosis | 1087 | Development only | Metrorrhagia | Conventional CM information | yes | RF; SVM; ANN; DT | NR |
| 18 | [14] | English | J Biomed Inform | Diagnosis | 570 | Development only | Diabetes | Tongue instrument parameters | NR | GA_XGBT; DT; KNN; LR; SVM; ANN; RF | NR |
| 19 | [14] | English | Int J Med Inform | Diagnosis | 1512 | Development with external validation | Diabetes | Conventional CM information | NR | NB; LR; RF; SVM; XGB; ANN; KNN; DT | NR |
| 20 | [25] | English | Comput Math Methods Med | Diagnosis | 342 | Development with external validation | CHD | Hand image parameters | NR | MTIALM | NR |
| 21 | [25] | English | J Integr Med | Diagnosis | 10060 | Development only | Liver cancer | Conventional CM information | yes | PSO-ELM; SVM; BN | NR |
| 22 | [32] | English | Biomed Pharmacother | Diagnosis | 586 | Development only | MS | Conventional CM information | NR | DT; SVM; RF | NR |
| 23 | [59] | Chinese | Doctoral dissertation | Diagnosis | 316 | Development with external validation | Hypertension | Pulse-taking instrument parameters | yes | RF; SVM; XGBoost; LGB;ANN | NR |
| 24 | [56] | Chinese | Masteral dissertation | Prognosis | 988 | Development with external validation | Liver cancer | Conventional CM information | yes | AdaBoost; NB; ANN; RF; SVM | NR |
| 25 | [47] | Chinese | Doctoral dissertation | Diagnosis | 1427 | Development only | Depression | Pulse-taking instrument parameters; Tongue instrument parameters | yes | DT;KNN;RF;Adaboost;GDBT;Bootstrap;NB;SVM | NR |
| 26 | [50] | Chinese | Masteral dissertation | Diagnosis | 610 | Development only | CHD | Pulse-taking instrument parameters; Tongue instrument parameters | yes | LR | NR |
| 27 | [53] | Chinese | Masteral dissertation | Prognosis | 103 | Development only | Lung cancer | Conventional tongue information | NR | LR | NR |
| 28 | [14] | Chinese | Chinese J of Basic Medicine of Chinese Medicine | Diagnosis | 852 | Development only | Diabetes | Conventional tongue information | NR | LR; AN; SVM; NB | NR |
| 29 | [65] | Chinese | Chinese J of CM | Diagnosis | 300 | Development with external validation | Infertility | Conventional CM information | yes | RF; SVM; KNN; ANN | NR |
| 30 | [55] | Chinese | Masteral dissertation | Diagnosis | 804 | Development only | Insomnia | Conventional CM information | yes | RF | NR |
| 31 | [4] | English | Frontiers in physiology | Diagnosis | 736 | Development only | Fatigue | Pulse-taking instrument parameters; Tongue instrument parameters | NR | LR; SVM; RF; ANN | NR |
| 32 | [21] | English | Comput Biol Med | Diagnosis | 1778 | Development with external validation | NAFLD | Conventional CM information | NR | AdaBoost; GBDT; NB; ANN; RF; SVM | NR |
| 33 | [4] | English | Biomed Res Int | Diagnosis | 522 | Development only | Lung cancer | Pulse-taking instrument parameters; Tongue instrument parameters | yes | RF; LR; SVM; ANN | NR |
| 34 | [27] | English | BMC Med Inform Decis Mak | Diagnosis | 12,000 | Development only | AIDS | Conventional CM information | yes | KNN; SVM; ANN; RF | NR |
| 35 | [24] | English | J Healthc Eng | Diagnosis | 950 | Development with external validation | Corporeity | Conventional CM information | yes | BPNN | NR |
| 36 | [21] | English | Medicine (Baltimore) | Diagnosis | 2436 | Development only | Gastropathy | Conventional CM information | yes | DT; SVM; RF | NR |
| 37 | [45] | Chinese | Masteral dissertation | Prognosis | 1713 | Development only | RA | Pulse-taking instrument parameters; Tongue instrument parameters | yes | KNN; SVM; DF; RF; ANN; AdaBoost | NR |
| 38 | [62] | Chinese | Chinese J of CM | Diagnosis | 622 | Development only | Diabetic Nephropathy | Modern CM instrument parameters | yes | ANN | NR |
| 39 | [30] | Chinese | Doctoral dissertation | Diagnosis | 1385 | Development only | CHD | Modern CM instrument parameters | yes | DF | NR |
| 40 | [48] | Chinese | Masteral dissertation | Diagnosis | 802 | Development only | Diabetes | Conventional CM information | NR | LR | NR |
| 41 | [38] | Chinese | Doctoral dissertation | Diagnosis | 708 | Development only | NAFLD | Tongue instrument parameters | NR | SVM | NR |
| 42 | [52] | Chinese | Masteral dissertation | Diagnosis | 340 | Development only | Stroke | Conventional CM information | yes | LR | NR |
| 43 | [28] | English | Evid Based Complement Alternat Med | Diagnosis | 523 | Development only | Hypertension | Pulse-taking instrument parameters | NR | LR | NR |
| 44 | [20] | English | JMIR Mhealth Uhealth | Diagnosis | 467 | Development only | Diabetes | Pulse-taking instrument parameters | NR | LR; RF; SVM | NR |
| 45 | [43] | Chinese | Masteral dissertation | Diagnosis | 733 | Development only | CHD | Conventional CM information | yes | SVM; ANN | NR |
| 46 | [36] | Chinese | Doctoral dissertation | Diagnosis | 515 | Development only | Lung cancer | Conventional tongue information | yes | AdaBoost; NB; ANN; RF; SVM | NR |
| 47 | [40] | Chinese | Doctoral dissertation | Diagnosis | 520 | Development only | Hypertension | Pulse-taking instrument parameters; Tongue instrument parameters | yes | Xgboost | NR |
| 48 | [35] | Chinese | J of Beijing University of CM | Diagnosis | 98 | Development only | Gastropathy | Conventional CM information | yes | RF; SVM | NR |
| 49 | [25] | English | Biomed Res Int | Diagnosis | 929 | Development only | Hypertension | Pulse-taking instrument parameters | NR | KNN; RF; AdaBoost; Gradient Boosting; SVM | NR |
| 50 | [41] | Chinese | Masteral dissertation | Diagnosis | 397 | Development only | Urticaria | Conventional tongue information | yes | ANN; DF | NR |
| 51 | [63] | Chinese | Information J of Chinese Medicine | Diagnosis | 919 | Development only | Gastropathy | Conventional CM information | yes | RF | NR |
| 52 | [34] | English | Biomed Res Int | Diagnosis | 827 | Development only | Diabetes | Tongue instrument parameters | NR | SVM; k-NN; NB; BPNN; GASVM | NR |
| 53 | [51] | Chinese | J China J of CM and Pharmacy | Diagnosis | 136 | Development only | Hepatic failure | Conventional CM information | NR | LR | NR |
| 54 | [26] | English | IEEE Trans Biomed Eng | Diagnosis | 525 | Development only | Corporeity | Conventional CM information | NR | RF; SVM; XGBoost; LGB; ANN | NR |
| AIDS: Acquired Immune Deficiency Syndromes; CHD: Coronary Heart Disease; MCI: Mild Cognitive Impairment; MS: Metabolic Syndrome; NAFLD: Non-Alcoholic Fatty Liver Disease; PCOS: Polycystic Ovarian Syndrome; RA: Rheumatoid Arthritis; ANN: Artificial Neutral Network; DT: Decision Tree; BPNN: Back Propagation Neural Network; DF; Extremely Randomized Trees, Extra trees; GDBT : Gradient Boosting; KNN: K Nearest Neighbor; LR: Logistic Regression; NB: Naive Bayes; SVM: Support Vector Machine; RF: Random Forest | |||||||||||
Reporting and methodological quality assessment
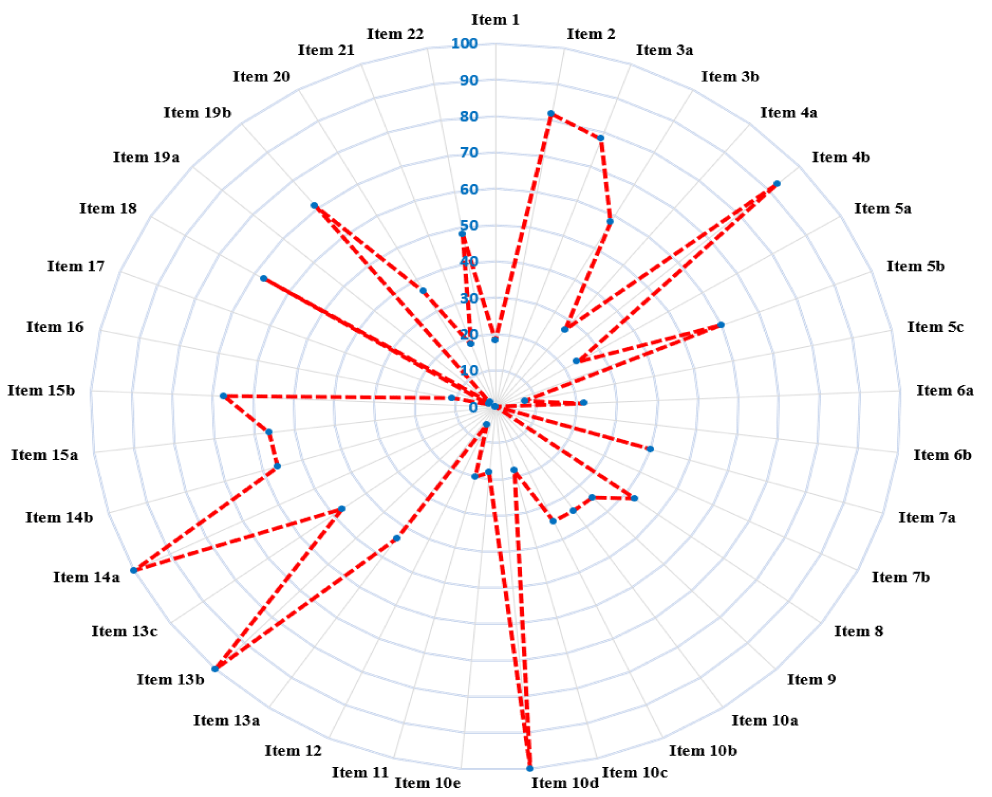
Based on the TRIPOD checklist5, the reporting quality of the CM diagnostic prediction studies was suboptimal. The results of the assessment are presented in figure 2 and further elucidated in Supplementary File 2-Data extraction. All primary studies (n = 54, 100%) reported sufficient details on item 10d (statistical analysis methods: specify all measures used to assess model performance), item 13b (participants: describe the characteristics of the participants), and item 14a (model development: specify the number of participants and outcome events in each analysis). Given the potential variation in abstract formats across different journals, the contents reported in the abstracts were evaluated rather than rigidly assessing item 2 against the "structured format" requirement. Then 50 (81.81%) studies reported sufficient details for this item. 14 studies (25.92%) reported sufficient details on item 4a (source of data: describe the study design or source of data) and (specify the key study dates). None of the studies achieved a "Y" rating for item 6b (Report any actions to blind assessment of the outcome to be predicted), and similarly, no study reported details on item 7b (report any actions to blind assessment of predictors for the outcome and other predictors). In terms of study size and missing data handling, 23 studies (42.59%) reported sufficient details on item 8 (explain how the study size was arrived at for predictors), and 15 (27.78%) described how missing data were handled (e.g., complete-case analysis, single imputation, multiple imputation). 10 included studies (18.52%) furnished performance measures with 95%Cls for the prediction model in item 16. Only 2 studies (3.7%) reported the results from model updating in item 17. Most of the studies reported study limitations in item 18 (n = 51, 94.44%), but only three studies (5.56%) discussed the results with regard to performance in the development data. 21 studies (38.89%) provided information about the availability of supplementary resources in item 21 (e.g., study protocol, web calculator, and data sets). 26 studies (48.15%) reported funding sources detailly in the study in item 22.
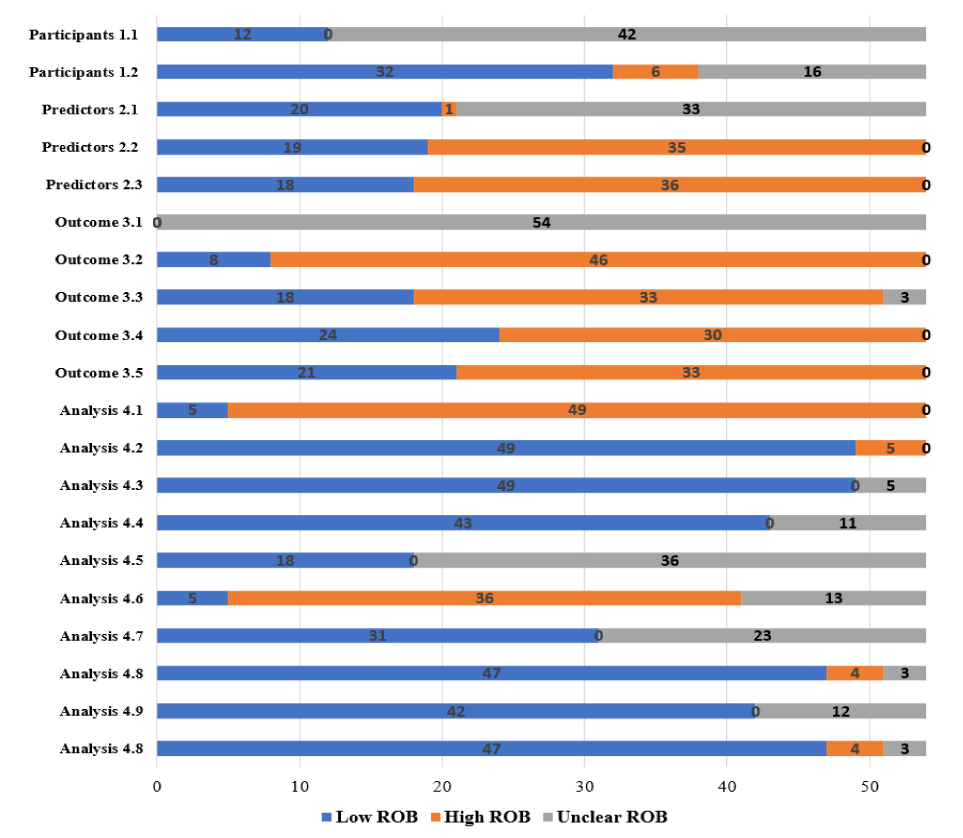
The methodological quality of the studies of CM diagnosis prediction model including four domains were evaluated by the PROBAST [6]. For domain 1: Participants, 13 studies (24.07%) reported study designs as a cohort design, nested case–control or case–cohort design (with proper adjustment of the baseline risk/hazard in the analysis), while the remains were unclear. 32 studies (59.26%) reported the inclusion and exclusion criteria of patients in the prediction model in detail and referred to the disease diagnostic criteria of Chinese and Western medicine, respectively. Six studies (11.11%) did not report CM diagnostic criteria in detail. There is no information on whether inappropriate inclusions or exclusions took place in 16 included studies (29.63%). For domain 2: Predictors, there is no information on how predictors were defined or assessed for most of the studies (n = 33, 61.11%). Meanwhile, most studies (n = 35, 64.81%) did not report detailed outcome information to evaluate the predictors and showed no information on whether predictors would be available at the time the model is intended to be used for prediction (n = 36, 66.67%). For domain 3: Outcome, the outcome determination has been used which is considered optimal or acceptable for all included studies (n = 54, 100%), while 46 of them mentioned but did not prespecified these methods (85.19%). 33 studies (61.11%) did not report information about whether the outcome was determined without knowing information about predictors. 49 studies (90.74%) did not provide information on time intervals to illustrate the relationship between predictor assessment and outcome ascertainment. For domain 4: Analysis, all studies reported information on the number of candidate predictor parameters or number of participants with the outcome, such that the Events Per Variable (EPV) can be calculated, while 10 of the studies them were not reported the reasonable number of participants (EPV < 10). Continuous and categorical predictors were handled appropriately in all primary studies (n = 54, 100%). 43 studies (79.63%) reported data analysis for all participants. The remains of 11 studies (20.37%) were with no information, in which five of the studies (9.26%) reported participants with missing data are omitted from the analysis. Although most studies (n = 47, 87.04%) appropriately evaluated relevant model performance metrics (e.g., calibration and discrimination) and had detailed internal validation techniques (e.g., bootstrapping and cross-validation). 45 studies (83.33%) did not provide on whether complexities (e.g., censoring, competing risks, sampling of control participants) in the data are present or accounted for appropriately. The details of the PROBAST assessment are presented in figure 3 and Supplementary File 2-Data extraction.
In addition, the correlation between the reporting and methodological quality of the included studies based on the number of"Y" and "low ROB" responses was moderate (rs= 0.504, p < 0.001). Sensitivity analysis did not decrease the correlation (rs= 0.568, p <0.001).
Extent of spin practices across sections
A thorough assessment of spin practices was conducted, categorizing 15 distinct spin practices that occurred across various sections: title, results, discussion, and conclusion in both the abstract and main text. Remarkably, no spin practices were identified in titles concerning inconsistency with study results or use of leading words (n = 0, 0%). Within the results section, three spin practices (n = 3, 5.56%) were observed, involving the use of leading words such as “Excellent” “significant”, “accurate”, and “improve”. Similar practices were also found in the discussion and conclusion section (n = 3, 5.56%). 27 studies (50%) and 43 studies (79.63%) used strong statements to describe the model in the abstract and the main text respectively. However, there is no spin practice in reporting differences between performance measures pre-specified in methods and results. In the main text, leading words were used as spin practices in 17 studies (31.48%) for results, 28 studies (51.85%) for discussion, and 28 studies (51.85%) for conclusion. Finally, qualifiers (such as ‘‘very’’ and ‘‘may’’) were used frequently to describe findings in the main text (n = 6, 11.11%). In the both of abstract and the main text, the diagnostic prediction model based on CM indicators, neither emphasized model relevance nor recommended application of the model in a different setting or population without external validation in the same study. Detailed information is presented in table 2 for further insights.
| Table 2: Frequency of ‘spin’ practices in abstract, and main text. | ||
| No. (%) | ||
| Abstract (n = 54) | Main text (n = 54) | |
| Title | ||
| Title is inconsistent with the study results | 0 (0) | NA |
| Use of leading words | 0 (0) | NA |
| Results section | ||
| Differences between performance measures pre-specified in Methods and reported in Results section | 0 (0) | 0 (0) |
| Use of leading words | 3 (5.56) | 17 (31.48) |
| Novel | 0 (0) | 3 (5.56) |
| Excellent | 1 (1.85) | 4 (7.41) |
| Accurate | 2 (3.7) | 0 (0) |
| Significant | 3 (5.56) | 16 (29.62) |
| Improved | 1(1.85) | 11 (20.37) |
| Use of strong statements to describe the model and/or model performance / accuracy / effectiveness | 27 (50) | 43 (79.63) |
| Spin in tables or figures | NA | 10 (18.52) |
| Non relevant models are not discussed | NA | 45 (83.33) |
| Discussion and conclusion section | ||
| Use of strong statements to describe the model and/or model performance / accuracy / effectiveness | 0 (0) | 13 (24.07) |
| Use of leading words | 3 (5.56) | 28 (51.85%) |
| Novel | 0 (0) | 3 (5.56) |
| Excellent | 1(1.85) | 4 (7.41) |
| Accurate | 2 (3.7) | 0 (0) |
| Significant | 3 (5.56) | 17 (31.48) |
| Improved | 1(1.85) | 23 (42.59) |
| Invalid comparison of results to previous development and/or validation studies is given | NA | 10 (18.52) |
| Nonrelevant models are not discussed | NA | 0 (0) |
| Emphasis on model relevance in the abstract while the results reported previously do not support such position | 0 (0) | 0 (0) |
| Discrepancy between full-text and abstract explanation of the study findings | 0 (0) | 0 (0) |
| Recommendation to use the model in clinical practice without external validation in same study | 2 (4.55) | 6 (13.63) |
| Recommendation to use the model in a different setting or population without external validation in the same study | 0 (0) | 7 (12.96) |
| Qualifiers are used | 6 (11.11) | 25 (42.30) |
| Very | 3 (5.56) | 19 (35.19) |
| May/Might | 6 (11.11) | 13 (24.07) |
| Other benefits not prespecified in Methods are addressed | NA | 0 (0) |
| Conclusions are inconsistent with the reported study results | 0 (0) | 0 (0) |
| Conclusion focuses solely on significant results | 6 (11.11) | 10 (18.52) |
Evidence mapping of CM diagnostic prediction model
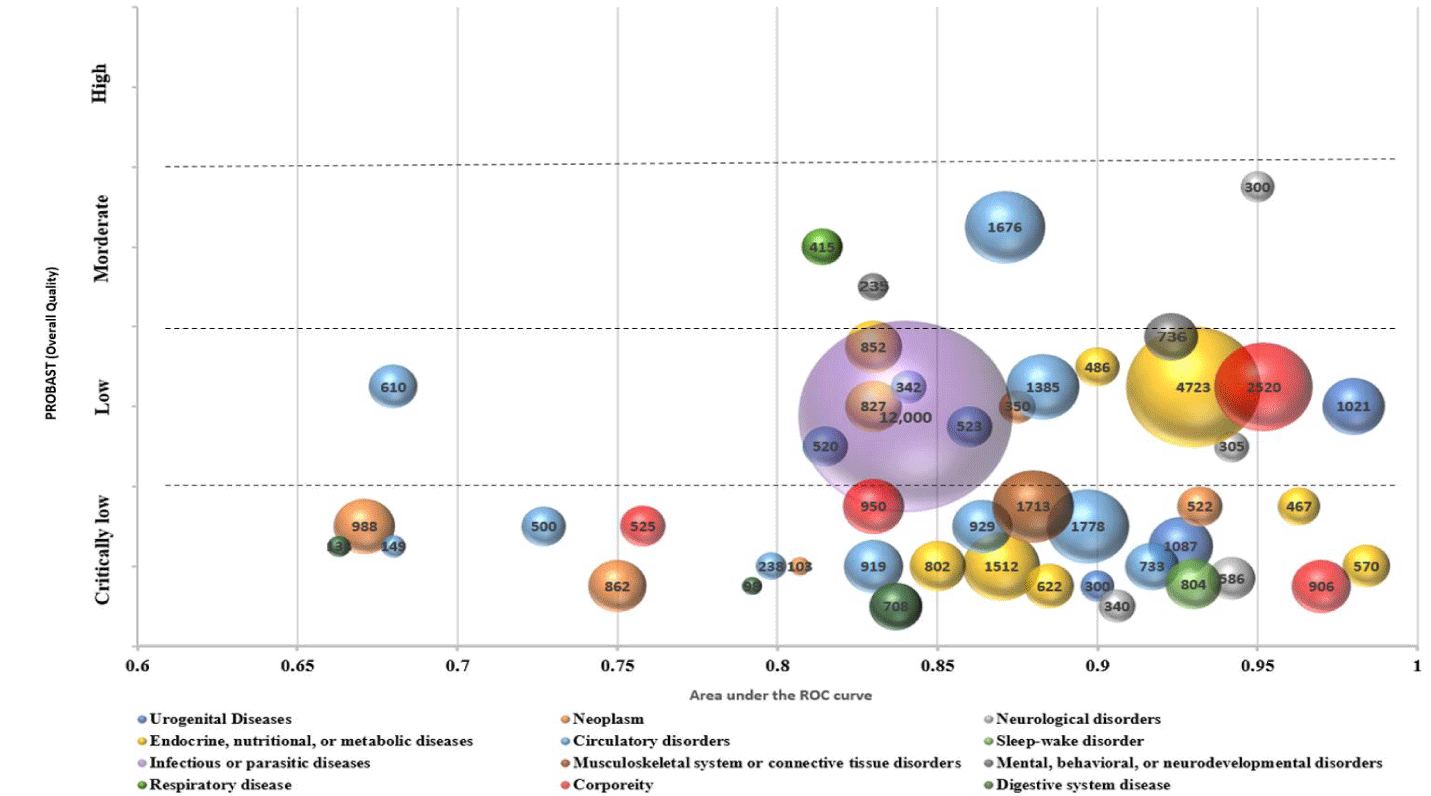
As shown in figure 4, the evaluation of overall methodological quality revealed that 36 CM diagnosis prediction model publications were judged as “Critically Low” quality, 14 studies were judged as “Low” quality, while only four were considered “Moderate” quality.
Based on ICD-11 categorization, the 54 included studies could be divided into 12 categories: Circulatory disorders (CHD: n = 5; hypertension: n = 6; cardiac failure: n = 1), Endocrine, nutritional, or metabolic diseases (PCOS: n = 1; diabetes: n=8), Neoplasm (lung cancer: n = 5; liver cancer: n = 2), Urogenital Diseases (metrorrhagia: n = 1; infertility: n = 1), Neurological disorders (Stroke: n = 3), Dermatologic Diseases (eczema: n = 1; urticaria: n = 1), Musculoskeletal system or connective tissue disorders (RA: n = 1), Infectious or parasitic diseases (AIDS: n = 1), Sleep-wake disorder (insomnia: n = 1), Mental, behavioral, or neurodevelopmental disorders (depression: n=1; fatigue: n = 1; MCI: n = 1), Digestive system disease (hepatic failure: n = 1; gastropathy: n = 2; NAFLD: n = 2), and corporeity (n = 4). Four studies with “Moderate” methodological quality were observed to predict COPD (AUC=0.814) [61], cardiac failure (AUC=0.871) [67], stroke (AUC = 0.95) [54], and MCI (AUC = 0.83) [60]. 15 disease prediction models diagnosed by CM yielded AUC values greater than 0.9. Among them, J Li, et al. [22] tested diabetics with the GA_XGBT model had reported the strongest predictive power, the AUC is 0.984 with the methodological quality judged as “Critically Low”. SJ Yao, et al. [66] evaluated menopause utilizing an RF model, achieving AUC of 0.98 with the methodological quality judged as "Low". In circulatory disorders studies, LL Hu reported the best decision tree predictive result for CHD (AUC=0.917) [43], with the quality of evidence was evaluated as “Critically Low”. YL Shi, et al. [30] demonstrated the Neural Network model provided the strongest predictive power for Non-Small-Cell Lung Cancer (NSCLC) based on objective CM indicators, achieving an AUC of 0.94, which also judged as "Critically Low" quality in terms of methodological quality.
Discussion
Principal findings
This systematic review is the first overview summarizing the clinical evidence for machine learning-based prediction models of CM diagnostic. Included studies mostly published in journals related to complementary medicine, intelligent computing, and biomed pharmacology from 2004 to 2023. The overall quality of reporting and methodological quality of this systematic review included in the present study were suboptimal, highlighting a pressing need for higher-quality publications in the realm of CM diagnostic prediction models. Content requiring marked improvement in reporting and method quality in terms of Title, details of participants, calculate of sample size, details of statistical analysis, model validation, details of model performance, interpretation, supplementary information, and funding report. For spin practice, attention needs to be paid for rigor of expression. And it is not recommended to apply models to different settings or populations without external validation in the same study.
Over recent years, a large number of CM prediction models have been published in the field of CM diagnostics, which usually provide risk estimates based on readily available clinical information, laboratory indicators, objective tongue and pulse parameters, etc. The integration of contemporary data science technology with traditional Chinese medicine diagnostic principles has yielded predictive CM diagnostic models capable of forecasting disease development and treatment outcomes for patients. With the reproducibility issues and the waste of research resources in biomedical studies have attracted considerable attention in the science community [70,71]. Researchers begin to know that the comprehensive and transparent reporting of the study design, the research process, and the final results are key safeguards to avoid the problems described above [72, 73]. As an existing review have observed, studies describing the development and validation (including updating) of prediction models often fail to report critical information, adherence to the TRIPOD checklist is strongly advocated [8]. Through open and transparent reporting of the evaluation process and results, the scientific validity and verifiability of CM diagnostics prediction model research can identify the problems and deficiencies of CM diagnostics prediction model [70]. We believed that the confidence of researchers in relative model research can be increased, the basis for further research can be provided. In addition, it should be noted that the assessment of methodological quality is based on the main content, so clear and comprehensive reporting is essential for the assessment of methodological quality6. As demonstrated in our study, we assessed the correlation between the reporting and methodological quality of the studies included in this study, similar to the article by Chapman, et al. [17], assessing the quality of systematic reviews in high-impact surgical journals. According to the results presented in this study, diagnostic criteria for CM and Western medicine need to be provided in detail to reduce selection bias in relevant studies of CM prediction models. The calculation of the sample size is essential. If the sample size cannot meet the calculated results, the algorithm of the predictive model needs to be improved to reduce the overfitting, which is the main effect due to the insufficient sample size. The selection of predictive models should be based on previously published studies, which can reduce the waste of resources. The detailed machine algorithm needs to be described in detail in the supplementary material. Most notably, external validation of prediction models is essential. Because external validation can show not only the stability of the predictive power, but also its generalization and generalization ability. This is the most important support for selecting to use for clinical.
According to the spin practice problems, we know the reward system within academia and the increasing amount of published research make spin in research, to some extent, necessary and therefore more frequent. As authors, our natural inclination is to ensure the publication of our work, leading us consciously and subconsciously to use the language to increase the credibility and readability of our findings [9]. A systematic review including 35 publications assessing misleading practices showed that spin evaluation varies per study design [10]. Navarro, et.al. reported that spin practices and poor reporting standards are also present in 152 studies, on prediction models using machine learning techniques9. We believe that a tailored framework for the identification of spin will enhance the sound reporting of prediction model studies. Therefore, this research not only added the evidence for CM model prediction of spin practice, as well as suggested that authors should make every effort to avoid distortion and hype in the further research.
The combination of four diagnostic methods in CM is partly used to realize the idea of "preventive treatment of diseases". Our results showed that there is a limited number of quantitative included models based on CM syndrome differentiation, as same as seldomly mentioned the content of complex traditional Chinese medicine diagnostic thinking process. Therefore, we hope that the incorporation of machine learning algorithms in CM diagnosis presents a viable approach to enhance the model performance for more discriminative classification of CM syndromes.
The evidence mapping results of this study showed that many of the predictive models based on CM diagnostic parameters had satisfactory test results, especially for diabetes, CHD, and lung cancer. However, the specific construction and optimization of the model still need the support of clinical data, and the clinical application value also needs further scientific evaluation. A number of studies have shown that the CM diagnosis prediction model based on tongue and pulse-taking instruments can accurately predict the development of patients' diseases and treatment effects [4,29,30]. However, this study has not yet further compared the prediction differences between the indicators based on conventional CM diagnosis and the objective prediction indicators of tongue diagnosis and pulse diagnosis parameters, as it has little relevance to the research topic of this review.
Strengths and limitations
To our knowledge, there has been no systematic review and evidence-mapping about spin practices and reporting quality in CM diagnosis studies and, particularly not in studies on machine learning-based prediction models. In terms of its advantages, the results of this study filled gaps in the knowledge and can be used to guide future research and clinical decision-making. Secondly, we prospectively registered the protocol on a widely recognized website, thus guaranteeing the transparency of our research process and avoiding the possibility of selection bias. Thirdly, we explored the relationship between the reporting and methodological quality of the included studies, and sensitivity analysis confirmed the robustness of the primary analysis. However, several limitations are worth highlighting. Firstly, we did not perform new meta-analyses by re-estimation of predicted value, because doing so would have been beyond the research aims of this research review. Secondly, we focused on the use of leading words in spin practices rather than allowing a certain degree of rhetoric and evaluating it within its specific context. Similarly, we could not determine if the use of qualifiers was detrimental because we only counted the occurrence rather than evaluating its use to show uncertainty. The assessment of spin practice inherently involves a degree of subjectivity, as reviewers' judgments play a pivotal role. While we diligently addressed any disagreements through discussion to minimize reading bias, it is plausible that others might interpret authors' statements differently, particularly in cases of linguistic spin. Thirdly, the absence of a comparison group limits our ability to establish causal relationships or infer associations between study characteristics, spin practices, and reporting standards. Our aim was to spotlight spin-indicative practices through a descriptive analysis, rather than delve into causal relationships. Despite these limitations, we still provided exploratory evidence about the presence of spin and reporting quality in CM diagnosis prediction model studies.
Conclusion
The available evidence indicates that predictive models based on CM indicators to predict diseases are worthy of consideration and can provide predictions for different diseases, but the scientific quality of published studies needs to be improved. Moreover, the predictive performance of CM diagnosis prediction models needs to be confirmed through high-quality external validation across multiple countries with large sample sizes. These concerted efforts are essential to solidify the credibility and applicability of predictive CM diagnosis models in real-world clinical scenarios.
Authors’ Contributions
Conceptualization, ZXX and JYL; methodology, JYL; software, JKL; validation, YM X and JKL; formal analysis, JYL; investigation, ZXX and JYL; resources, JYL; data curation, JKL and JYL; writing—original draft preparation, JYL; writing—review and editing, JYL and ZXX; visualization, JKL and JYL; supervision, ZXX, XXA and MZ; All authors have read and agreed to the published version of the manuscript.
Data availability
The Ethics Committee of Shanghai University of Traditional Chinese Medicine limited the measurement data used to support the results of this study in order to protect the privacy of patients. For researchers who meet the criteria for obtaining confidential data, the data of this study are available from the corresponding author upon request.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Funding
This research was funded by the National Natural Science Foundation of China (No.82074333) and Shanghai Key Laboratory of Health Identification and Assessment (No.21DZ2271000).
References
- Li Z, Ren X, Xiao L, Qi J, Fu T, Li W. Research on Data Analysis Network of TCM Tongue Diagnosis Based on Deep Learning Technology. J Healthc Eng. 2022 Mar 29;2022:9372807. doi: 10.1155/2022/9372807. PMID: 35392154; PMCID: PMC8983244.
- Li G, Guo Y. Exploring the Medication Pattern of Chinese Medicine for Peptic Ulcer Based on Data Mining. J Healthc Eng. 2021 Nov 11;2021:9072172. doi: 10.1155/2021/9072172. Retraction in: J Healthc Eng. 2022 Nov 29;2022:9782953. PMID: 34804461; PMCID: PMC8601794.
- Wang W, Zeng W, He S, Shi Y, Chen X, Liping Tu, Yang B, Xu J, Yin X. A new model for predicting the occurrence of polycystic ovary syndrome: based on data of tongue and pulse. Digital health 2023;9:20552076231160323. doi/10.1177/20552076231160323.
- Shi YL, Liu JY, Hu XJ, Tu LP, Cui J, Li J, Bi ZJ, Li JC, Xu L, Xu JT. A New Method for Syndrome Classification of Non-Small-Cell Lung Cancer Based on Data of Tongue and Pulse with Machine Learning. Biomed Res Int. 2021 Aug 11;2021:1337558. doi: 10.1155/2021/1337558. PMID: 34423031; PMCID: PMC8373490.
- Collins GS, Reitsma JB, Altman DG, Moons KG. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): the TRIPOD statement. BMJ. 2015 Jan 7;350:g7594. doi: 10.1136/bmj.g7594. PMID: 25569120.
- Wolff RF, Moons KGM, Riley RD, Whiting PF, Westwood M, Collins GS, Reitsma JB, Kleijnen J, Mallett S; PROBAST Group†. PROBAST: A Tool to Assess the Risk of Bias and Applicability of Prediction Model Studies. Ann Intern Med. 2019 Jan 1;170(1):51-58. doi: 10.7326/M18-1376. PMID: 30596875.
- Li Q, Chi L, Zhao W, et al. Machine learning prediction of motor function in chronic stroke patients: a systematic review and meta-analysis. Frontiers in neurology 2023;14:1039794.
- Andaur Navarro CL, Damen JAA, Takada T, Nijman SWJ, Dhiman P, Ma J, Collins GS, Bajpai R, Riley RD, Moons KGM, Hooft L. Systematic review finds "spin" practices and poor reporting standards in studies on machine learning-based prediction models. J Clin Epidemiol. 2023 Jun;158:99-110. doi: 10.1016/j.jclinepi.2023.03.024. Epub 2023 Apr 5. PMID: 37024020.
- Sensever FA, de Lucena Alves CP, Lima GDS, Loomans B, Opdam N, Pereira-Cenci T. Spin and reporting in systematic reviews with meta-analysis of randomized clinical trials in restorative dentistry. J Dent. 2022 Oct;125:104282. doi: 10.1016/j.jdent.2022.104282. Epub 2022 Sep 7. PMID: 36084762.
- Chiu K, Grundy Q, Bero L. 'Spin' in published biomedical literature: A methodological systematic review. PLoS Biol. 2017 Sep 11;15(9):e2002173. doi: 10.1371/journal.pbio.2002173. PMID: 28892482; PMCID: PMC5593172.
- Page MJ, McKenzie JE, Bossuyt PM, Boutron I, Hoffmann TC, Mulrow CD, Shamseer L, Tetzlaff JM, Akl EA, Brennan SE, Chou R, Glanville J, Grimshaw JM, Hróbjartsson A, Lalu MM, Li T, Loder EW, Mayo-Wilson E, McDonald S, McGuinness LA, Stewart LA, Thomas J, Tricco AC, Welch VA, Whiting P, Moher D. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. BMJ. 2021 Mar 29;372:n71. doi: 10.1136/bmj.n71. PMID: 33782057; PMCID: PMC8005924.
- Cumpston M, Li T, Page MJ, Chandler J, Welch VA, Higgins JP, Thomas J. Updated guidance for trusted systematic reviews: a new edition of the Cochrane Handbook for Systematic Reviews of Interventions. Cochrane Database Syst Rev. 2019 Oct 3;10(10):ED000142. doi: 10.1002/14651858.ED000142. PMID: 31643080; PMCID: PMC10284251.
- Kempf E, de Beyer JA, Cook J, Holmes J, Mohammed S, Nguyên TL, Simera I, Trivella M, Altman DG, Hopewell S, Moons KGM, Porcher R, Reitsma JB, Sauerbrei W, Collins GS. Overinterpretation and misreporting of prognostic factor studies in oncology: a systematic review. Br J Cancer. 2018 Nov;119(10):1288-1296. doi: 10.1038/s41416-018-0305-5. Epub 2018 Oct 24. PMID: 30353050; PMCID: PMC6251031.
- Lu C, Ke L, Li J, Zhao H, Lu T, Mentis AFA, Wang Y, Wang Z, Polissiou MG, Tang L, Tang H, Yang K. Saffron (Crocus sativus L.) and health outcomes: a meta-research review of meta-analyses and an evidence mapping study. Phytomedicine. 2021 Oct;91:153699. doi: 10.1016/j.phymed.2021.153699. Epub 2021 Aug 8. PMID: 34419735.
- Lu C, Ke L, Li J, Wu S, Feng L, Wang Y, Mentis AFA, Xu P, Zhao X, Yang K. Chinese Medicine as an Adjunctive Treatment for Gastric Cancer: Methodological Investigation of meta-Analyses and Evidence Map. Front Pharmacol. 2022 Jan 10;12:797753. doi: 10.3389/fphar.2021.797753. PMID: 35082677; PMCID: PMC8784830.
- Harrison JE, Weber S, Jakob R, Chute CG. ICD-11: an international classification of diseases for the twenty-first century. BMC Med Inform Decis Mak. 2021 Nov 9;21(Suppl 6):206. doi: 10.1186/s12911-021-01534-6. PMID: 34753471; PMCID: PMC8577172.
- Chapman SJ, Drake TM, Bolton WS, Barnard J, Bhangu A. Longitudinal analysis of reporting and quality of systematic reviews in high-impact surgical journals. Br J Surg. 2017 Feb;104(3):198-204. doi: 10.1002/bjs.10423. Epub 2016 Dec 21. PMID: 28001294.
- Chen X, Wang Y, Zhang L, Xu X. Construction and Evaluation of Neural Network Correlation Model between Syndrome Elements and Physical and Chemical Indexes of Unstable Angina Pectoris Complicated with Anxiety. Comput Math Methods Med. 2022 Aug 21;2022:6217186. doi: 10.1155/2022/6217186. Retraction in: Comput Math Methods Med. 2023 Oct 4;2023:9807987. PMID: 36045947; PMCID: PMC9420622.
- Ding L, Zhang XY, Wu DY, Liu ML. Application of an extreme learning machine network with particle swarm optimization in syndrome classification of primary liver cancer. J Integr Med. 2021 Sep;19(5):395-407. doi: 10.1016/j.joim.2021.08.001. Epub 2021 Aug 4. PMID: 34462241.
- Hao Y, Cheng F, Pham M, Rein H, Patel D, Fang Y, Feng Y, Yan J, Song X, Yan H, Wang Y. A Noninvasive, Economical, and Instant-Result Method to Diagnose and Monitor Type 2 Diabetes Using Pulse Wave: Case-Control Study. JMIR Mhealth Uhealth. 2019 Apr 23;7(4):e11959. doi: 10.2196/11959. PMID: 31012863; PMCID: PMC6658300.
- Jiang T, Guo XJ, Tu LP, Lu Z, Cui J, Xiang Ma X, juan Hu X, Hua Yao X, Tao Cui L, zhi Li Y, Bin Huang J, Xu J. Application of computer tongue image analysis technology in the diagnosis of nafld. Computers in biology and medicine 2021;135:104622. doi: 10.1016/j.compbiomed.2021.104622.
- Li J, Chen Q, Hu X, Yuan P, Cui L, Tu L, Cui J, Huang J, Jiang T, Ma X, Yao X, Zhou C, Lu H, Xu J. Establishment of noninvasive diabetes risk prediction model based on tongue features and machine learning techniques. Int J Med Inform. 2021 May;149:104429. doi: 10.1016/j.ijmedinf.2021.104429. Epub 2021 Feb 22. PMID: 33647600.
- Li J, Yuan P, Hu X, Huang J, Cui L, Cui J, Ma X, Jiang T, Yao X, Li J, Shi Y, Bi Z, Wang Y, Fu H, Wang J, Lin Y, Pai C, Guo X, Zhou C, Tu L, Xu J. A tongue features fusion approach to predicting prediabetes and diabetes with machine learning. J Biomed Inform. 2021 Mar;115:103693. doi: 10.1016/j.jbi.2021.103693. Epub 2021 Feb 1. PMID: 33540076.
- Luo Y, Lin B, Zhao S, He L, Wen C. Neural Network-Based Study about Correlation Model between TCM Constitution and Physical Examination Indexes Based on 950 Physical Examinees. J Healthc Eng. 2020 Sep 1;2020:8812678. doi: 10.1155/2020/8812678. PMID: 32952990; PMCID: PMC7481996.
- Luo ZY, Cui J, Hu XJ, Tu LP, Liu HD, Jiao W, Zeng LZ, Jing CC, Qiao LJ, Ma XX, Wang Y, Wang J, Pai CH, Qi Z, Zhang ZF, Xu JT. A Study of Machine-Learning Classifiers for Hypertension Based on Radial Pulse Wave. Biomed Res Int. 2018 Nov 11;2018:2964816. doi: 10.1155/2018/2964816. PMID: 30534557; PMCID: PMC6252205.
- Pang B, Zhang D, Li N, Wang K. Computerized tongue diagnosis based on Bayesian networks. IEEE Trans Biomed Eng. 2004 Oct;51(10):1803-10. doi: 10.1109/TBME.2004.831534. PMID: 15490827.
- Pang H, Wei S, Zhao Y, He L, Wang J, Liu B, Zhao Y. Effective attention-based network for syndrome differentiation of AIDS. BMC Med Inform Decis Mak. 2020 Oct 15;20(1):264. doi: 10.1186/s12911-020-01249-0. PMID: 33059709; PMCID: PMC7558604.
- Qi Z, Zhao ZY, Xu JT, et al. Radial Pulse Wave Signals Combined with Ba-PWV for the Risk Prediction of Hypertension and the Monitoring of Its Accompanying Metabolic Risk Factors. Evidence-based complementary and alternative medicine : eCAM 2020;2020:3926851.
- Shi Y, Guo D, Chun Y, Liu J, Liu L, Tu L, Xu J. A lung cancer risk warning model based on tongue images. Front Physiol. 2023 Jun 1;14:1154294. doi: 10.3389/fphys.2023.1154294. PMID: 37324390; PMCID: PMC10267397.
- Shi Y, Yao X, Xu J, Hu X, Tu L, Lan F, Cui J, Cui L, Huang J, Li J, Bi Z, Li J. A New Approach of Fatigue Classification Based on Data of Tongue and Pulse With Machine Learning. Front Physiol. 2022 Feb 7;12:708742. doi: 10.3389/fphys.2021.708742. PMID: 35197858; PMCID: PMC8859319.
- Wang Q, Zhao C, Qiang Y, Zhao Z, Song K, Luo S. Multitask Interactive Attention Learning Model Based on Hand Images for Assisting Chinese Medicine in Predicting Myocardial Infarction. Comput Math Methods Med. 2021 Oct 26;2021:6046184. doi: 10.1155/2021/6046184. PMID: 34737789; PMCID: PMC8563122.
- Xia SJ, Gao BZ, Wang SH, Guttery DS, Li CD, Zhang YD. Modeling of diagnosis for metabolic syndrome by integrating symptoms into physiochemical indexes. Biomed Pharmacother. 2021 May;137:111367. doi: 10.1016/j.biopha.2021.111367. Epub 2021 Feb 13. PMID: 33588265.
- Ye H, Gao Y, Zhang Y, Cao Y, Zhao L, Wen L, Wen C. Study on intelligent syndrome differentiation neural network model of stomachache in traditional Chinese medicine based on the real world. Medicine (Baltimore). 2020 May 29;99(22):e20316. doi: 10.1097/MD.0000000000020316. PMID: 32481403.
- Zhang J, Xu J, Hu X, Chen Q, Tu L, Huang J Cui J. Diagnostic method of diabetes based on support vector machine and tongue images. BioMed research international 2017;2017:7961494. doi: 10.1155/2017/7961494.
- Y Cao, Y Lu, JX Chen, et al. Application of traditional Chinese medicine intelligent syndrome differentiation model for gastroesophageal reflux disease based on machine learning. Journal of Beijing University of TCM, 2019;42(10).
- LZ Zeng. Study on diagnostic features and risk model of tongue image in patients with non-small cell lung cancer. Shanghai University of TCM, Doctoral dissertation. 2019.
- C Chen. Study on early warning model of coronary heart disease risk event based on four diagnostic parameters of traditional Chinese medicine. Shanghai University of TCM, Doctoral dissertation. 2020.
- JL Chen. Establishment of NAFLD prediction model based on tongue diagnosis and correlation study of liver fat content by magnetic resonance imaging. Academy of Chinese Medical Sciences, Doctoral dissertation. 2020.
- ML Chen. The tongue image characteristics of lung cancer patients based on digital tongue map. Beijing University of TCM, Masteral dissertation.2022.
- RB Chen. Research on diagnosis and prediction model of hyperliver-fire syndrome in hypertension based on XGboost algorithm. Academy of Chinese Medical Sciences, Doctoral dissertation. 2018.
- YN Chu. Construction of prediction model and classifier of urticaria syndrome type based on ANN. Guangzhou University of TCM, Masteral dissertation. 2018.
- . HR Cui. Research on classification model of TCM syndrome of disintegrating leakage based on machine learning. Changchun University of TCM, Masteral dissertation. 2018.
- LL Hu. To study the risk prediction model of coronary heart disease based on the information of four diagnostic methods of traditional chinese medicine. Shanghai University of TCM, Masteral dissertation. 2022.
- Jun Li, XJ Hu, LP Tu, et al. A type 2 diabetes risk prediction model based on tongue image parameters combined with multiple index features. Chinese Journal of Basic Medicine of Chinese Medicine, 2021;27(03):451-56+501.
- Y Li. Using feature selection and machine learning algorithm to predict TCM syndrome type of patients with rheumatoid arthritis. University of Science and Technology of China, Masteral dissertation. 2020.
- Y Li. Clinical prediction model of coronary heart disease combined with chronic heart failure. Guangzhou University of TCM, Masteral dissertation. 2021.
- JF Lin. To establish a traditional chinese medicine artificial intelligence syndrome differentiation scheme for depression based on a variety of physiological information and symptoms and signs. Shanghai University of TCM, 2021. Doctoral dissertation.
- YT Lin. Analysis of the characteristics of tongue and pulse diagnosis in patients with abnormal blood glucose. Shanghai University of TCM, Masteral dissertation. 2020.
- JT Liu. Construction of clinical prediction model of hypertension with left ventricular hypertrophy combined with traditional chinese and western medicine. Tianjing University of TCM, 2020. Masteral dissertation.
- Y Liu, P Yuan, X Qi, et al. Study on the diagnostic value of traditional Chinese medicine pulse chart parameters for coronary heart disease in hypertensive patients. Tianjin traditional Chinese medicine, 2022;39(08):982-86.
- QJ Ru, WJ Ye, DH Yang, et al. Quantitative assessment of gastric gas in patients with liver failure and its relationship with prognosis. J China Journal of Traditional Chinese Medicine and Pharmacy 2012;27(5):1304-08.
- BC Song. Analysis of risk factors and establishment of clinical prediction model for vascular dementia (deficiency stasis syndrome). Beijing University of TCM, Masteral dissertation. 2020.
- JR Sun. Correlation study and prediction of tongue image characteristics and myelosuppression after chemotherapy in lung cancer. Beijing University of TCM, Masteral dissertation. 2021.
- ZJ Sun, J J, CY Ma, et al. Journal of Hunan University of Chinese Medicine. 2023;43(4).
- Tang Y, Li Z, Yang D, Fang Y, Gao S, Liang S, Liu T. Research of insomnia on traditional Chinese medicine diagnosis and treatment based on machine learning. Chin Med. 2021 Jan 6;16(1):2. doi: 10.1186/s13020-020-00409-8. PMID: 33407711; PMCID: PMC7789502.
- HP Wang. To study the clinical prediction model of survival prognosis of colorectal cancer liver metastasis involving tcm factors. Academy of Chinese Medical Sciences, Masteral dissertation. 2021.
- HW Wang. Construction of diabetic foot risk prediction model based on tongue image. J Trad Chin Med. 2022;63(19).
- Q Wang. Exploration of establishing and verifying the diagnosis model of phlegm-stasis syndrome of apoplexy based on lasso regression. J Journal of Zhejiang Chinese Medical University 2023;47(4):416-21.
- XR Wang. A preliminary study of artificial intelligence methodology for predicting blood pressure and virtual demonstration based on three-channel pulse information. Beijing University of TCM, Doctoral dissertation. 2021.
- Feng W. Clinical prediction model of mci phlegm turbidification blocking oqiao syndrome and clinical study on treating mci phlegm turbidification blocking oqiao syndrome with qianxin decoction. Hubei University of TCM, Doctoral dissertation. 2022.
- Y Wu. Establishment of clinical prediction model of COPD complicated with chronic pulmonary heart disease and study on distribution of TCM syndromes. Tianjin University of TCM, 2022. Masteral dissertation.
- TW Xia. Based on machine learning, the prediction model of chinese and western multi-modal feature fusion of type 2 diabetes complicated with kidney disease was constructed. Chengdu University of TCM, Doctoral dissertation. 2020.
- WW Xu, WJ Gu, GP Liu, et al. Empirical feature selection and syndrome classification recognition of chronic gastritis based on random forest and multi-label learning algorithm. Information Journal of Chinese Medicine. 2016;23(08):18-23.
- YQ Xu, JD Yang, Q Jiang, et al. Research on tongue image classification of tcm syndrome based on multi-feature fusion. Intelligent computers and applications. 2022;12(07):25-34.
- MB Xu, YF Liu, ZY Zhao, et al. Construction and application of infertility tcm syndrome differentiation model based on artificial intelligence. Chinese Journal of Traditional Chinese Medicine. 2021;36(09):5532-36.
- CF Du, JH Yang, Y Yao. Construction of chinese medicine intelligent diagnosis model of perimenopausal syndrome based on integrated learning. J Trad Chin Med 2023;64(6).
- Y Yang. Development of clinical prediction model for hospital death of heart failure by combination of chinese and western medicine. Liaoning University of TCM, Doctoral dissertation. 2022.
- Y Zhang. Research on prediction model of TCM syndrome type of eczema based on machine learning. Liaoning University of TCM, Masteral dissertation. 2022.
- Y Zhao, YS Li, XR Wu. Crack tongue recognition algorithm based on tongue image multi-feature fusion and machine learning. Journal of Yanshan University. 2022;46(06):522-28.
- Damen JA, Hooft L, Schuit E, Debray TP, Collins GS, Tzoulaki I, Lassale CM, Siontis GC, Chiocchia V, Roberts C, Schlüssel MM, Gerry S, Black JA, Heus P, van der Schouw YT, Peelen LM, Moons KG. Prediction models for cardiovascular disease risk in the general population: systematic review. BMJ. 2016 May 16;353:i2416. doi: 10.1136/bmj.i2416. PMID: 27184143; PMCID: PMC4868251.
- Glasziou P, Altman DG, Bossuyt P, Boutron I, Clarke M, Julious S, Michie S, Moher D, Wager E. Reducing waste from incomplete or unusable reports of biomedical research. Lancet. 2014 Jan 18;383(9913):267-76. doi: 10.1016/S0140-6736(13)62228-X. Epub 2014 Jan 8. PMID: 24411647.
- Jiang T, Hu XJ, Yao XH, Tu LP, Huang JB, Ma XX, Cui J, Wu QF, Xu JT. Tongue image quality assessment based on a deep convolutional neural network. BMC Med Inform Decis Mak. 2021 May 5;21(1):147. doi: 10.1186/s12911-021-01508-8. PMID: 33952228; PMCID: PMC8097848.
- Chalmers I, Glasziou P. Avoidable waste in the production and reporting of research evidence. Lancet. 2009 Jul 4;374(9683):86-9. doi: 10.1016/S0140-6736(09)60329-9. Epub 2009 Jun 12. PMID: 19525005.
Content Alerts
SignUp to our
Content alerts.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.