Biology Group. 2024 February 03;5(2):106-116. doi: 10.37871/jbres1877.
One-Year Investigation of Hepatitis A Virus and Rotavirus in Two Waste Water Treatment Plants in Egypt
Sahar Abd Al-Daim*
- Waste water treatment plant
- Rotavirus
- Hepatitis A virus
- Sludge
- Inlet, outlet
Abstract
Background: Waterborne enteric viruses are evolving source of disease outbreaks and represent a major threat to global public health. The repetitive assessment of water environments is necessary to manage enteric virus-mediated fecal infection and the probable emergence of different variants.
Methods: Here, we detected human rotavirus and hepatitis A virus circulating in two wastewater treatment plants, for one year in sewage and sludge samples from two largest wastewater treatment plant (El-Gabal El-Asfar and Zenin) located in Egypt. For this purpose, we examine predefined viruses' prevalence in 72 samples (24 raw sewage samples, 24 treated effluent samples, and 24 sludge) collected from August 2022 to July 2023. Viral RNA was extracted and detected by nested Reverse Transcriptase-Polymerase Chain Reaction (RT-PCR) in case of HAV and by multiplex semi nested RT-PCR In case of rotavirus with the
Results: Percentage of detection rate 58.3%, 25% 41.6%, 50%, 16.6%58.6%, 41.6%, 8.3%, 41.6%41.6%, 8.3% and 41.6% for inlet, outlet, sludge in WWTP zenin and WWTP El-Gabal El-Asfar for HAV, RV respectively. Using generic primer result indicated G1P8 was the most common although G2P [4], G1, G2, G3P? G9P [8], G9P [11], G1 P [4] were detected with different percentage.
Conclusion: These results demonstrated that HAV and RV still remain in the environment after sewage treatment and could play an important role in maintaining the endemicity of HAV and RV infection. Routine and continuous monitoring of waterborne viruses are most useful approach to understand disease occurrence in communities.
Graphical abstract
Abbreviations
RV: Rotavirus; HAV: Hepatitis A Virus; WWTP: Waste Water Treatment Plant; RT: Reverses Transcriptase; PCR: Polymerase Chain Reaction; VP7: Viral Protein 7; VP4:Viral Protein; RNA: Ribo Nuclic Acid; CSOs: Combined Sewer Overflows
Introduction
A large number of epidemics are being caused by waterborne enteric viruses, which pose a serious risk to public health worldwide. Enteric viruses can spread quickly in aquatic ecosystems with low removal rate, and they can come from human wastes [1]. Therefore, appropriate risk management requires surveillance and source apportionment of enteric viruses in ambient waters. Every year, there are over 4 billion episodes of diarrheal illnesses caused by wastewater, and 2 million people die from them, the majority of them are younger than five years old [1]. The most significant way of transmission for enteric viruses is through direct contact with infected people. Enteric viruses are spread through the fecal-oral pathway [2].
Hepatitis A (HAV) is one of the main aetiologic mediators of acute hepatitis worldwide, [3,4], is a 27–32 nm, single-stranded RNA virus with no envelope and positive sense with a genome of 7.5 kbp [5]. HAV has seven genotypes and one serotype. Globally, the epidemiology of HAV is evolving in a number of geographic areas. Over 100 million HAV infections are thought to occur annually in the world [6]. The existence of HAV in sewage has been thoroughly investigated and well-documented [3,7]. While the frequency of acute HAV infections and the seroprevalence of hepatitis A have decreased in Europe, waste Water-Based Epidemiology (WBE) has improved our understanding of the epidemiological changes in disease trends [8]. Thus, Different European nations have reported varying frequencies of HAV detection in urban wastewater, ranging from 3.1% to 60% [8-11]. Countries with poorer resources and more endemicity, such as South Africa, Egypt, and Tunisia, have shown higher detection frequencies (>60%) [12-14].
RV are major causes of acute diarrhea in infants, leading to 9% mortality in the under-5 age group globally [15]. The genome of RV consists of 11 segments that make up double-stranded RNA (dsRNA). One of the six structural viral proteins (VP1-4, VP6, and VP7) or five to six non-structural proteins (NSP1–5/6) are encoded by each segment [16]. Belong to Reoviridae family. Can survive at a wide pH range [3-11] and for at least 7, 14, 22 and 32 days at 37 °C, 20 °C, 4 °C and − 20 °C, respectively with a slight loss of infectivity at 37 °C after 4 days [17,18]. The high RV incidence rate imposes a significant burden on both low- and high-income countries [19]. RV infection results in approximately 39% of diarrheal-mediated mortality, with a larger proportion occurring in developing countries [6].
In low-income countries, a lot of areas lack adequate sanitary infrastructure and wastewater treatment facilities, hence fecal matter contaminates the environment and drinking water sources [20]. Additionally, large volumes of untreated wastewater may also be discharged via Combined Sewer Overflows (CSOs) during heavy rainfall events and via dry water overflows for example during snowmelt, tidal infiltration or system failures and blockages [21]. Enteric viruses tend to adsorb to solid particles in the form of sludge, and they are easily spread in environmental waters [22].
Because wastewater is frequently used for irrigation in countries where freshwater is scarce, enteric viruses having the potential to directly contaminate fruit and salad vegetables, leading to foodborne/water borne outbreaks [23,24].
To investigate pathogen abundance, persistence, adsorption, and movement in the aquatic environment, continuous evaluation of wastewater is required. This study aimed to perform monitoring of HAV and RV for one year in sewage and sludge samples from two large wastewater treatment plant (El-Gabal El-Asfar and Zenin) and to assess environmental contamination and its dissemination after treatment by an activated sludge process.
Materials and Methods
Study area
This study was conducted in two waste water treatment plants located in Greater Cairo governorate which is the capital city of Egypt, El-Gabal El-Asfar and Zenin WWTPs use an activated sludge as a treatment technology and the flow rate in El-Gabal El-Asfar was 1,700,000 cubic meters per day (m3 / day) while the flow rate in Zenin was 330,000 m3 /day [25]. El-Gabal El-Asfar receives raw sewage of a large area in Cairo Governorate and Zenin receives raw sewage of a large area in El-Giza Governorate which considered part of greater Cairo (Figure 1).
Samples collection
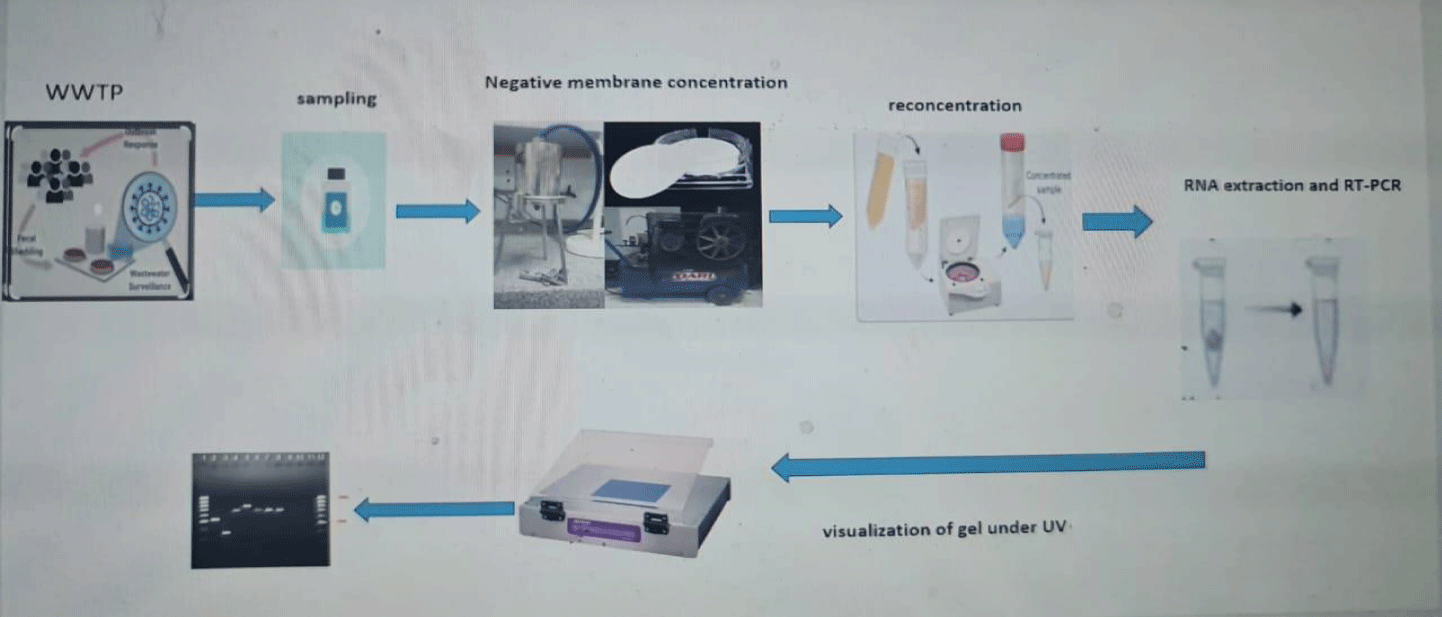
Two liters of each raw sewage (inlet), treated sewage (outlet) and sludge samples (100 grams) were collected from two waste water treatment plant (El-Gabal El-Asfarr WWTP and Zenin WWTP). Seventy-two waste water samples were collected monthly from both WWTPs from August 2022 to July 2023. Samples were immediately stored at 4 °C and transported in a cooler with ice packs to the laboratory in clean plastic bottles where processing began within 24h of collection.
Pre-analytical processing
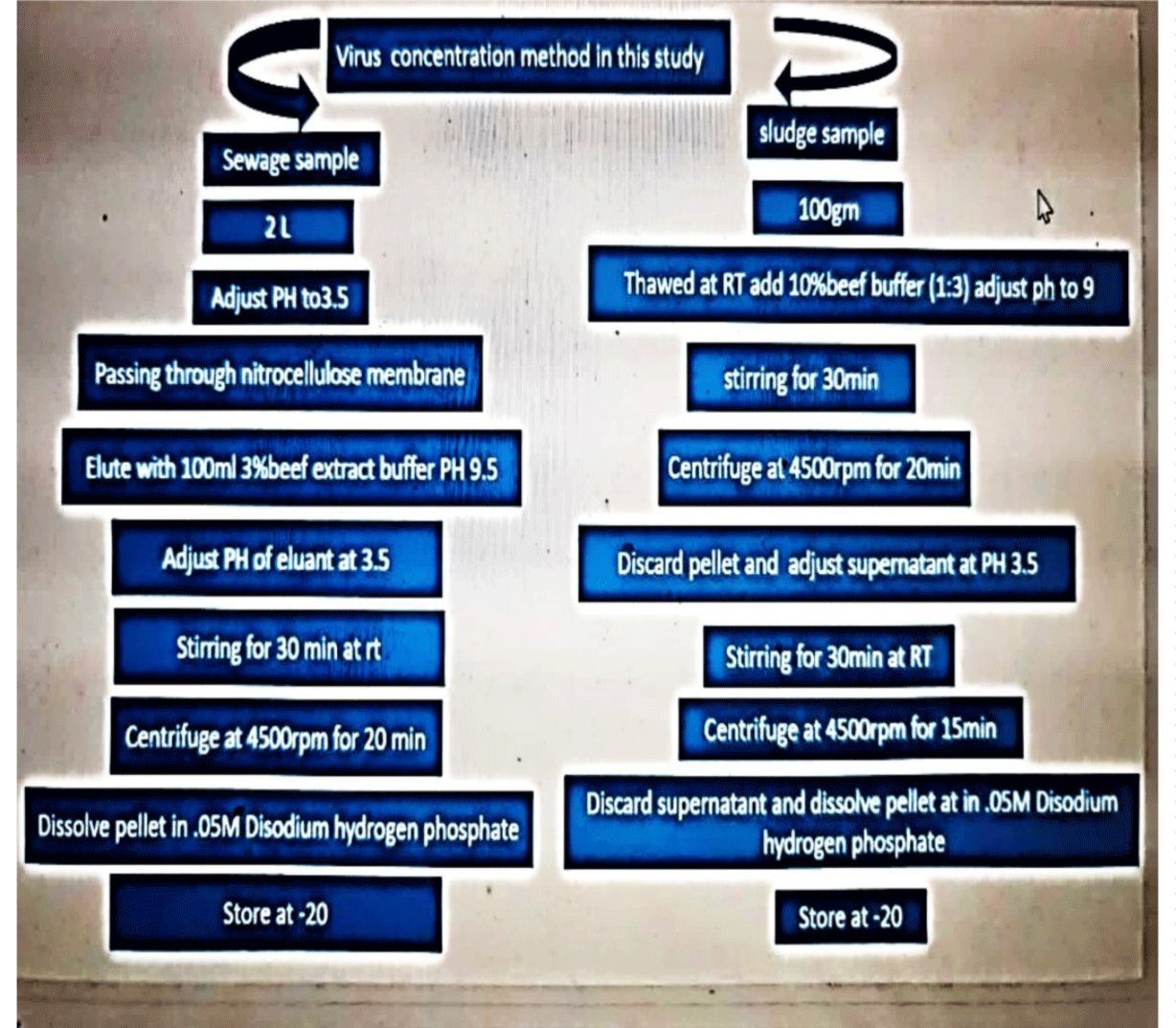
Virus concentration from inlet and outlet samples: An adsorption-elution method using nitrocellulose membrane was used to concentrate viruses from sewage samples as described previously with minor modification [26]. Briefly, sewage sample (2 L) was adjusted to PH 3.5 using 1M HCL and the sample was passed through stainless holder under pressure. Although, MgCl2 (2.5 mol l−1) was added to the sample to a final concentration of 0.05 mol l−1, and the pH was adjusted to 3.5 by hydrochloric acid (0.5 mol l−1). After filtration through a nitrocellulose cellulose membrane filter (0.45 μm, ADVANTEC, Tokyo, Japan). The viruses were eluted from the membranes, using 100 ml of a 3 % beef extract glycine solution (pH = 9), followed by organic flocculation precipitation procedure [27]. The eluted solution was stirred for 30 min in magnetic stirrer and then PH was adjusted to 3.5 and centrifuged at 4500 rpm for 20 min, then the supernatant was discarded and the pellet were dissolved in 2ml of 0.05M disodium hydrogen phosphate solution and stored at -20 until being used in RNA extraction. The obtained eluent was treated with antibiotic antimitotic to kill any bacteria or fungus present in the sample.
Virus concentration from sludge samples: One hundred gram of sludge sample were weighted and then thawed by stirring at room temperature then 300 ml (1g of sample: 3 g of buffer) (10%beef extract buffer). The mixture stirred for 30 min at PH 7 and centrifuged at 4500 rpm for 15min then obtained pellet were discarded and obtained supernatant was adjusted at PH 3.5 stirred for 30min, centrifuged at 4500 rpm for 15min, finally the obtained pellet was dissolved in 0 .15 M Na2HPO4, stored at -80 until analyzed by RT-PCR [26]. The obtained eluent was treated with antibiotic antimitotic to kill any bacteria or fungus present in the sample (Figure 2).
Molecular detection
Genomic RNA was extracted from 140 μL of the eluate to obtain a final volume of 60 μL, using the QIAamp Viral RNA (Qiagen, Germany) according to the manufacturer’s instructions. The RNA genome for both RV and HAV were converted to cDNA using first strand cDNA synthesis protocol (heat block) this conversion is necessary because that cDNA is a more convenient way to work with the coding than RNA, which could be degraded easily by RNases present in the samples. The extracted RNA and cDNA were stored in -20 until used in PCR. cDNA was carried out in heating block of the DNA-thermal cycler (Bio-Rad, France) for 5min to allow opening the helices of the RNA template.
cDNA synthesis for HAV and RV: cDNA was synthesized by mixing 5 µl of the extracted RNA, 2 µl of specific reverse primer for each virus, which were heated at 65OC for 5min to allow opening the helices of the RNA template then chilled on ice for 2 min to prevent formation of secondary structural. Then we add 2 µl of dNTP, 0.5μl of Maxima Reverse Transcriptase (Thermo scientific (200 U/μl)), 5µl of 5x RT buffer, 0.5µl RNase inhibitor, 10 µl DEPC-treated water to make 25 µl reaction mixtures.
The mixture was heated at 30°C for 30 min, 42°C for 1 hr., followed by 95°C for 5 min, to stop working enzyme and then chilled on ice. In case of Rotavirus RNA was subjected to denaturation at 65°C for 15 min and then chill on ice for 5 min (Table 1).
| Table 1: Primer sequence used for HAV detection. | ||||
| PCR round | Primer sequence | Orientation | band size | Reference |
| 1st | A-F1 5’-CTATTCAGATTGCAAATTAYAAT-3’ A-R1 5’-AAYTTCATYATTTCATGCTCCT-3’ |
Sense Antisense |
391bp | [27] |
| 2nd | A-F2 5’-TATTTGTCTGTYACAGAACAATCAG-3’ A-R2 5’-AGGRGGTGGAAGYACTTCATTTGA-3’ |
Sense antisense |
244bp | |
Nested RT-PCR for HAV: The polymerase chain reaction used to detect HAV were consisted of two rounds of PCR in the first round we used 3ul of synthesized cDNA and 12.5ul of Dream Taq PCR master mix ,1ul of forward primer A-F1,1ul of A-R1reverse primer and 7.5ul of DEPEC water to obtain reaction mixture of 25ul with temp condition of 5min at 95c followed by 40 cycles of denaturation 94c for 45sec, annealing 55c for 45 sec and extension 72c for 1min, with final extension at 72c for 10min and the obtained PCR product were under go to second round of PCR condition with another two specific primer with 1ul of PCR product of first round and 12.5ul of dream Taq master mix ,1ul of forward primer A-F2 , 1ul of reverse primer A-R2 and 9.5 ul of DEPEC water with PCR condition as follow : primary denaturation 95c for 5min, 35cycles of 94c fo30 sec ,62c for 45sec and 72c for 1min with final extension time at72c for 5min.
Semi nested multiples RT-PCR for RV: Oligonucleotide primer sequences were obtained from Applied Biosynthesis Company in Germany, as purified lyophilized primers for amplification of VP7 and VP4 outer capsid protein of RV. PCR was carried out using 5µl of the synthesized cDNA in 0.2 ml DNase/RNase free Eppendorf tube, were added to reaction volume of 25µl: 12.5 Dream Taq master mix, 1µl of each sense (VP7-F) and antisense primer (VP7-R), 9.5 µl DEPC water in case of G typing, con3 antisense primer and con2 sense primer in case P typing in which each of which is consisted of semi nested multiplex typing primers as in the tables 2,3. The PCR mixture was subjected to the temperature condition as in table 4.
| Table 2: Primer sequence for G typing of RV. > | ||||
| Primer> | Sequence (5-3)> | Ampli (bp)> | Reference> | |
| 1st> | VP7-F > | ATG TAT GGT ATT GAA TAT ACC AC > | 881 > | [28] > |
| 2nd> | VP7-R > | AAC TTG CCA CCA TTT TTT CC > | ||
| > | G3 > | ACG AAC TCA ACA CGA GAG G > | 682 > | [29] > |
| G9 > | CTT GAT GTG ACT AYa A AAT AC > | 179 > | ||
| G10 > | ATG TCA GAC TAC ARb A TAC TGG > | 266 > | ||
| G8 > | GTC ACA CCA TTT GTA AAT TCG > | 754 > | [30] > | |
| G4 > | CGT TTC TGG TGA GGA GTT G > | 452 > | ||
| G2 > | CAA TGA TAT TAA CAC ATT TTC TGT G > | 521 > | ||
| G1 > | CAA GTA CTC AAA TCA ATG ATG G > | 618 > | ||
| Table 3: Primer sequence for P typing of RV. > | ||||
| Primer> | Sequence (5-3)> | Amplicon (bp)> | Reference> | |
| 1st > | Con3 > | TGGCTTCGCCATTTLATAGACA > | 876 > | [31] > |
| Con2 > | ATTTCGGACCAT'lTATAACC > | |||
| 2nd > | Rota p8 > | TCTACTTGGRTTRACNTGC > | 345 > | [32] > |
| 2T-1 > | CTATTGTTAGAGGTTAGAGTC > | 483 > | [33] > | |
| 3T-1 > | TGTTGATTAGTTGGATTCAA > | 267 > | ||
| 4T-1 > | TGAGACATGCAATTGGAC > | 391 > | ||
| 5T-1 > | ATCATAGTTAGTAGTCGG > | 583 > | ||
| Rotap11 > | GTAAACATCCAGAATGTG> | 312 > | [34] > | |
| Table 4: PCR Condition for First and Second Round for Rotavirus. | ||||||||
| PCR steps and conditions | Denaturation | Annealing | Extension | No. cycles | ||||
| Temp (°C) | Time (min) | Temp (°C) | Time (min) | Temp (°C) | Time (min) | |||
| G typing | 1st round | 95 | 1 | 52 | 1 | 72 | 1 | [35] |
| 2nd round | 95 | 1 | 42 | 2 | 72 | 1 | [30] | |
| P typing | 1st round | 95 | 1 | 50 | 2 | 72 | 1 | [35] |
| 2nd round | 95 | 1 | 45 | 2 | 72 | 1 | [30] | |
Multiplex semi-nested RT-PCR for G typing of Rotaviruses: PCR typing could also be performed from PCR that was obtained from the first amplification of the entire gene 9. In this case, 2 µl of the PCR product served as the template for second amplification for typing. The same reaction buffer was used, but the primer mix containing all six serotype-specific primers G1, G2, G3, G4, G8, G9 and G10 and the common primer VP7-R were used. This primer sequence was shown in table 2. The PCR program was as mentioned in table 4 with final extension time at 72°C for 10 min.
Multiplex semi-nested RT-PCR for P typing of Rotaviruses: The typing primers were selected from the regions of gene 4 known to be highly divergent between strains in different genetic groups and highly conserved in strain from the same group. Six specific typing primers were prepared for gene 4 (3T-1, 2T-1, 4T-1, 5T-1, Rota p8, Rotap11) table 3.
For PCR experiments, dsRNA was prepared from sewage samples. Two amplification procedure was usually used. For first amplification, the cDNA was added to reaction mixture containing 3ul, 1 µl con3, 1 µl con 2, and 12.5ul of dream Taq master mix and 7.5ul DEPEC water. The tubes were placed in a thermocycler (Bio-Rad) and temperature condition as mentioned in table 4 and a final 10-min incubation at 72°C.
PCR products of 10 µl were mixed with 2 µl loading dye and analyzed by electrophoresis (General biosystem, Germany) using 1.5% agarose gel (electrophoresis grade, iNtRoN, Cat.No.32033) containing 0.5µg ethidium bromide. The PCR product was run at 100 V for 30 min in submarine electrophoresis (model: HB1214, RATED: 0-150V,0-100ma), and visualized under transilluminator (SPECTROLINE, MODEL-TM-312A), electrophoresis power supply (CONSORT,3000v-300Ma, E833) comparing to the low-grade DNA ladder (NORGEN, cat# 11400, Canda).
Results
Rotavirus was detected 50%, 16.6%, 58.6%, 41.6%, 8.3% and 41.6% in inlet, outlet, sludge (zenin WWTP), inlet, outlet, sludge (EL-Gabal EL-Asfar WWTP) with high percentage of detection in inlet and sludge samples. HAV was detected in 58.3%, 25%, 41.6%, 41.6%, 33.3% and 50% in inlet, outlet, sludge (zenin WWTP), inlet, outlet, sludge (EL-Gabal EL-Asfar WWTP) with high percentage of detection in inlet and sludge samples, which means raw sewage and sludge contain large number of viruses than treated sewage due to treatment process in two WWTPS mentioned in our study, with 33.3% virus removal in zenin and 8.3% virus removal in EL-Gabal EL-Asfar for HAV and 33.4% virus removal in zenin, 33.3%virus removal rate for RV. It was observed codetection of RV and HAV in inlet samples (zenin WWTP) in three months November, December and March although in two months of sludge samples (January and February). In El-Gabal El-Asfar WWTP it was observed codetection in three months one in inlet samples and two in sludge, April, October, and March respectively. With detection of rotavirus year around with high detection rate in color months, while HAV detected throughout the year with no peak in any season.
By using generic multiplex primer, we observed that the most common genotypes detected in our study is G1 P [8] followed by G2P [4], G1, G2, G3P? G9P [8], G9P [11], and finally G1 P [4] respectively with percentage of detection rate as shown in table 7 (Tables 5,6).
| Table 5: Detection of rotavirus and hepatitis A virus in different type of WWTP. | ||||||
| ZeninWWTP | El-Gabal El-AsfarrWWTP | |||||
| P/T | P/T | P/T | P/T | P/T | P/T | |
| Type of virus | inlet | outlet | sludge | Inlet | outlet | sludge |
| HAV | 7/12 | 3/12 | 5/12 | 5/12 | 4/12 | 6/12 |
| RV | 6/12 | 2/12 | 7/12 | 5/12 | 1/12 | 5/12 |
| Table 6: Distribution of rotavirus and hepatitis A virus throughout the year. | ||
| Months/year | Zenin WWTP total/ positive HAV RV |
El-Gabal El-Asfar WWTP total/ positive HAV RV |
| August 2022 | 2/3 0/3 | 1/3 0/3 |
| September 2022 | 1/3 0/3 | 1/3 0/3 |
| October 2022 | 0/3 2/3 | 1/3 1/3 |
| November 2022 | 1/3 3/3 | 2/3 1/3 |
| December 2022 | 2/3 1/3 | 0/3 3/3 |
| January 2023 | 1/3 3/3 | 1/3 1/3 |
| February 2023 | 1/3 1/3 | 1/3 1/3 |
| March 2023 | 2/3 2/3 | 2/3 2/3 |
| April 2023 | 0/3 2/3 | 1/3 2/3 |
| May 2023 | 2/3 1/3 | 0/3 0/3 |
| June 2023 | 1/3 0/3 | 3/3 0/3 |
| July2023 | 2/3 0/3 | 2/3 0/3 |
| Positive/total | 15/36 15/36 | 15/36 11/36 |
| percentages | 41.6% 41.6% | 41.6% 30.5% |
| Table 7: G and P types of rotaviruses detected in the present study. | |
| G/ P types | % of detection |
| G1 P8 | 38.4% |
| G2 P4 | 23.2% |
| G1, G3 P? | 15.5% |
| G9 P8 | 11.5% |
| G9 P11 | 7.6% |
| G1 P4 | 3.8% |
Discussion
Accurate risk management requires surveillance of enteric viruses in environmental waste waters. Approximately 80% of infections reported globally are thought to be waterborne [34-36]. The degree of wastewater contamination depends on the prevalence of viral infections and characteristics of viruses circulating in a given population [37]. Wastewater-based epidemiology uses concentrations of infectious disease targets in wastewater to understand disease occurrence in communities because wastewater represents a composite biological sample that contains contributions from every individual using drains and toilets in the sewer shed. Human excretions including feces, saliva, urine, blood, and mucus all enter the wastewater system. When individuals are infected with a pathogen, they excrete the pathogen (infectious or non-infectious) via excretions that contribute to wastewater. All people, regardless of disease status, contribute to wastewater, even though the quantity of pathogens shed by an individual may vary depending on the degree or course of their sickness, as well as whether they have symptoms or not. This suggests that a low-bias method of figuring out how diseases arise in communities is wastewater monitoring. Information gleaned from wastewater monitoring can testing availability and test-seeking behaviors of individuals [38,39].
Prior to virus identification, environmental samples are frequently concentrated for the precise detection of low viral titers. Water sample concentration is frequently achieved using ultracentrifugation, ultrafiltration, adsorption/elution, and flocculation; their advantages and disadvantages have already been discussed [40]. The kind of material, the virus type, and the concentration method all affect how well a virus may be recovered. In the present study, the method used was the negative charge concentration method (nitrocellulose) followed by the second organic flocculation concentration procedure because electronegative filters have comparatively good recoveries for frequently tested enteroviruses and are widely available, inexpensive, and of low cost.
Viral detection was detected in all samples in the current investigation by employing nested and semi-nested RT-PCR amplification, which increased amplification efficiency, decreased false-positive results, and improved detection rate. The prevalence, genotypes, seasonality, and prevalence of RV, also presence of HAV RNA in the greater Cairo, Egypt, all examined in this article. El-Gabal El-Asfar and Zenin, two sizable WWTPs, were tracked monthly for a year. RNA was still present in treated samples even though our results indicated a decrease in its detection. The public health is more likely to become infected with RV-A as a result of this outcome if agricultural water uses this wastewater again or if it is released into swimming and recreational water. Our study detects RV RNA totally with 42.6%which is considered as the same detection rate of sewage water in France,42% and considered Higher than study carried out in Egypt, 2019 with detection rate of 29.4% and lower than study carried out in Tunisia and China with percentage of detection 72.4, 93.5 respectively [41-43]. Other researchers in Egypt detect RV in waste water ranging from 21.9% to 50% [44-46].
Following several documented cases of HAV outbreaks among travelers from Europe, Egypt was deemed to be an endemic nation [47,48] because of high incidence of prevalence of HAV [49]. According to previous study carried out in Egypt, 83.3% of the wastewater samples taken from Zenin contained HAV [50]. Which is higher than our detection rate 41.6% indicating a sharp decrease in HAV spreading which may be attribute sanitary condition improved after pandemic, and although different time of collection. It was observed that HAV were found around the year with no seasonal variation.
Fifteen sewage and sludge samples that tested positive for rotavirus were genotyped using multiplex semi nested RT-PCR. G1P8 is most common genotypes in our study which is in agreement with studies carried out in Spain, Brazil, Venezuela [51-53].
According to the current study, there is a greater incidence of RV-A detection in the winter. We have found the similar seasonality pattern on clinical surveillances [41,45]. It is still unclear why viruses are more common in the winter.
Conclusion
G1 and P [8] strains of group A rotavirus are the most common in the environment, according to our study's characterization of the VP7 and VP4 genes from sewage samples is good method, to known the circulated and most common strain circulated in our population, to take it in consideration when development of specific vaccine. For this reason, our study suggests that environmental surveillance of sewage provides a good assessment of group A rotavirus genotypes circulating in the local human community. In order to gather valuable information for epidemiological purposes and outbreak early warning, and provide a useful data on the epidemiology of enteric virus infections circulating in the population, including asymptomatic infections future research should look at the incidence of other enterically transmitted viruses and incorporate both clinical and environmental samples with wide range of collected area and samples number.
Acknowledgement
We are grateful to the worker of waste water treatment plant who helped to collect the specimens used in this study.
Ethical approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Conflict of interests
No conflict of interest.
Funding
This research received no external funding.
Authors' contributions
S. Abd Al-Daim, Conceptualization, methodology, investigation, Writing-original draft preparation, reviewing, sampling.
Authors' information
Sahar Abd Al-Daim, Ph.D., Researcher of Virology, Environmental Virology Lab, Water Pollution Research Department, Environment and Climate Change Institute, National Research Center.
References
- Nicole SU, Garry AL, Caroline K. Occurrence of Human Enteric Viruses in Water Sources and Shellfish: A Focus on Africa, Food and Environmental Virology. 2021;13:1-31. doi: 10.1007/s12560-020-09456-8.
- Kitajima M, Gerba CP. Aichi virus 1: environmental occurrence and behavior. Pathogens. 2015 May 19;4(2):256-68. doi: 10.3390/pathogens4020256. PMID: 25996404; PMCID: PMC4493473.
- Global Hepatitis Report Geneva. WHO. 2017.
- Lemon SM, Ott JJ, Van Damme P, Shouval D. Type A viral hepatitis: A summary and update on the molecular virology, epidemiology, pathogenesis and prevention. J Hepatol. 2017 Sep 5:S0168-8278(17)32278-X. doi: 10.1016/j.jhep.2017.08.034. Epub ahead of print. PMID: 28887164.
- Aboubakr H, Goyal S. Involvement of Egyptian Foods in Foodborne Viral Illnesses: The Burden on Public Health and Related Environmental Risk Factors: An Overview. Food Environ Virol. 2019 Dec;11(4):315-339. doi: 10.1007/s12560-019-09406-z. Epub 2019 Sep 27. PMID: 31560123.
- Progress on Household Drinking Water, Sanitation and Hygiene 2000-2017. Special Focus on Inequalities. United Nations Children’s Fund (UNICEF) and World Health Organization. New York. 2019.
- Taylor MB, Cox N, Vrey MA, Grabow WO. The occurrence of hepatitis A and astroviruses in selected river and dam waters in South Africa. Water Res. 2001 Aug;35(11):2653-60. doi: 10.1016/s0043-1354(00)00551-0. PMID: 11456164.
- Bivins A, North D, Ahmad A, Ahmed W, Alm E, Been F, Bhattacharya P, Bijlsma L, Boehm AB, Brown J, Buttiglieri G, Calabro V, Carducci A, Castiglioni S, Cetecioglu Gurol Z, Chakraborty S, Costa F, Curcio S, de Los Reyes FL 3rd, Delgado Vela J, Farkas K, Fernandez-Casi X, Gerba C, Gerrity D, Girones R, Gonzalez R, Haramoto E, Harris A, Holden PA, Islam MT, Jones DL, Kasprzyk-Hordern B, Kitajima M, Kotlarz N, Kumar M, Kuroda K, La Rosa G, Malpei F, Mautus M, McLellan SL, Medema G, Meschke JS, Mueller J, Newton RJ, Nilsson D, Noble RT, van Nuijs A, Peccia J, Perkins TA, Pickering AJ, Rose J, Sanchez G, Smith A, Stadler L, Stauber C, Thomas K, van der Voorn T, Wigginton K, Zhu K, Bibby K. Wastewater-Based Epidemiology: Global Collaborative to Maximize Contributions in the Fight Against COVID-19. Environ Sci Technol. 2020 Jul 7;54(13):7754-7757. doi: 10.1021/acs.est.0c02388. Epub 2020 Jun 12. PMID: 32530639.
- Kamel AH, Ali MA, El-Nady HG, Deraz A, Aho S, Pothier P, Belliot G. Presence of enteric hepatitis viruses in the sewage and population of Greater Cairo. Clin Microbiol Infect. 2011 Aug;17(8):1182-5. doi: 10.1111/j.1469-0691.2011.03461.x. Epub 2011 Mar 7. PMID: 21375654.
- Rodrígue, RA, Pepper ILA, Gerba CP. Application of PCR-based methods to assess the infectivity of enteric viruses in environmental samples. Applied and Environmental Microbiology. 2009;75(2):297-307. doi: 10.1128/AEM.01150-08.
- Petrignani M, Verhoef L, de Graaf M, Richardus JH, Koopmans M. Chronic sequelae and severe complications of norovirus infection: A systematic review of literature. J Clin Virol. 2018 Aug;105:1-10. doi: 10.1016/j.jcv.2018.05.004. Epub 2018 May 24. PMID: 29804008.
- Pintó RM, Alegre D, Domínguez A, El-Senousy WM, Sánchez G, Villena C, Costafreda MI, Aragonès L, Bosch A. Hepatitis A virus in urban sewage from two Mediterranean countries. Epidemiol Infect. 2007 Feb;135(2):270-3. doi: 10.1017/S0950268806006753. Epub 2006 Jul 3. PMID: 16817987; PMCID: PMC2870566.
- Rachida S, Matsapola PN, Wolfaardt M, Taylor MB. Genetic characterization of a novel hepatitis a virus strain in irrigation water in South Africa. J Med Virol. 2016 Apr;88(4):734-7. doi: 10.1002/jmv.24370. Epub 2015 Sep 21. PMID: 26331799.
- Ouardani I, Manso CF, Aouni M, Romalde JL. Efficiency of hepatitis A virus removal in six sewage treatment plants from central Tunisia. Appl Microbiol Biotechnol. 2015 Dec;99(24):10759-69. doi: 10.1007/s00253-015-6902-9. Epub 2015 Aug 19. PMID: 26286509.
- Goel AK, Chawla S, Dhingra A, Thiyagarajan V, Nair NP. Rotavirus Diarrhea and its Determinants Among Under-Five Children Admitted in a Tertiary Care Hospital of Southern Haryana, India. Indian J Pediatr. 2021 Mar;88(Suppl 1):16-21. doi: 10.1007/s12098-020-03616-1. Epub 2021 Jan 27. PMID: 33501607.
- Greenberg HB, Estes MK. Rotaviruses: from pathogenesis to vaccination. Gastroenterology. 2009 May;136(6):1939-51. doi: 10.1053/j.gastro.2009.02.076. Epub 2009 May 7. PMID: 19457420; PMCID: PMC3690811.
- Pinon A, Vialette M. Survival of Viruses in Water. Intervirology. 2018;61(5):214-222. doi: 10.1159/000484899. Epub 2018 Jan 10. PMID: 29316545.
- Meng ZD, Birch C, Heath R, Gust I. Physicochemical stability and inactivation of human and simian rotaviruses. Appl Environ Microbiol. 1987 Apr;53(4):727-30. doi: 10.1128/aem.53.4.727-730.1987. PMID: 3034154; PMCID: PMC203745.
- Dbaibo G, Rajab M, Inati A, Mikhael R, Choueiry E, Al-Tannir M, Salam O, Ramakrishnan G, DeAntonio R. Hospital-based surveillance study of rotavirus gastroenteritis in children under 5 years of age in Lebanon. Trials Vaccinol. 2013;2:25-30. doi: 10.1016/j.trivac.2013.08.002.
- Bányai K, Estes MK, Martella V, Parashar UD. Viral gastroenteritis. Lancet. 2018 Jul 14;392(10142):175-186. doi: 10.1016/S0140-6736(18)31128-0. Epub 2018 Jun 29. PMID: 30025810; PMCID: PMC8883799.
- Ahmed W, Payyappat S, Cassidy M, Harrison N, Besley C. Sewage-associated marker genes illustrate the impact of wet weather overflows and dry weather leakage in urban estuarine waters of Sydney, Australia. Sci Total Environ. 2020 Feb 25;705:135390. doi: 10.1016/j.scitotenv.2019.135390. Epub 2019 Nov 23. PMID: 31838427.
- Hassard F, Gwyther CL, Farkas K, Andrews A, Jones V, Cox B, Brett H, Jones DL, McDonald JE, Malham SK. Abundance and Distribution of Enteric Bacteria and Viruses in Coastal and Estuarine Sediments-a Review. Front Microbiol. 2016 Nov 1;7:1692. doi: 10.3389/fmicb.2016.01692. PMID: 27847499; PMCID: PMC5088438.
- Chigor VN, Sibanda T, Okoh AI. Assessment of the risks for human health of adenoviruses, hepatitis A virus, rotaviruses and enteroviruses in the Buffalo River and three source water dams in the Eastern Cape. Food Environ Virol. 2014 Jun;6(2):87-98. doi: 10.1007/s12560-014-9138-4. Epub 2014 Mar 28. PMID: 24676673.
- Iritani N, Kaida A, Abe N, Kubo H, Sekiguchi J, Yamamoto SP, Goto K, Tanaka T, Noda M. Detection and genetic characterization of human enteric viruses in oyster-associated gastroenteritis outbreaks between 2001 and 2012 in Osaka City, Japan. J Med Virol. 2014 Dec;86(12):2019-25. doi: 10.1002/jmv.23883. Epub 2014 Jan 10. PMID: 24415518.
- Latif EF. Applying novel methods in conventional activated sludge plants to treat low-strength wastewater. Environmental Monitoring and Assessment. 2022;194:323. doi: 10.1007/s10661-022-09968-9.
- Rose JB, Singh SN, Gerba CP, Kelley LM. Comparison of microporous filters for concentration of viruses from wastewater. Appl Environ Microbiol. 1984 May;47(5):989-92. doi: 10.1128/aem.47.5.989-992.1984. PMID: 6742838; PMCID: PMC240036.
- Katzenelson E, Fattal B, Hostovesky T. Organic flocculation: an efficient second-step concentration method for the detection of viruses in tap water. Appl Environ Microbiol. 1976 Oct;32(4):638-9. doi: 10.1128/aem.32.4.638-639.1976. PMID: 10841; PMCID: PMC170320.
- Villar LM, Morais LM, Aloise R, Melo MM, Calado IA, Lampe E, Gaspar AM. Co-circulation of genotypes IA and IB of hepatitis A virus in Northeast Brazil. Braz J Med Biol Res. 2006 Jul;39(7):873-81. doi: 10.1590/s0100-879x2006000700004. PMID: 16862277.
- Iturriza-Gómara M, Isherwood B, Desselberger U, Gray J. Reassortment in vivo: driving force for diversity of human rotavirus strains isolated in the United Kingdom between 1995 and 1999. J Virol. 2001 Apr;75(8):3696-705. doi: 10.1128/JVI.75.8.3696-3705.2001. PMID: 11264359; PMCID: PMC114861.
- Iturriza-Gómara M, Kang G, Gray J. Rotavirus genotyping: keeping up with an evolving population of human rotaviruses. J Clin Virol. 2004 Dec;31(4):259-65. doi: 10.1016/j.jcv.2004.04.009. PMID: 15494266.
- Gouvea V, Glass RI, Woods P, Taniguchi K, Clark HF, Forrester B, Fang ZY. Polymerase chain reaction amplification and typing of rotavirus nucleic acid from stool specimens. J Clin Microbiol. 1990 Feb;28(2):276-82. doi: 10.1128/jcm.28.2.276-282.1990. PMID: 2155916; PMCID: PMC269590.
- Gentsch JR, Glass RI, Woods P, Gouvea V, Gorziglia M, Flores J, Das BK, Bhan MK. Identification of group A rotavirus gene 4 types by polymerase chain reaction. J Clin Microbiol. 1992 Jun;30(6):1365-73. doi: 10.1128/jcm.30.6.1365-1373.1992. PMID: 1320625; PMCID: PMC265294.
- Gómara MI, Cubitt D, Desselberger U, Gray J. Amino acid substitution within the VP7 protein of G2 rotavirus strains associated with failure to serotype. J Clin Microbiol. 2001 Oct;39(10):3796-8. doi: 10.1128/JCM.39.10.3796-3798.2001. PMID: 11574622; PMCID: PMC88438.
- Malik A, Yasar A, Tabinda A, Abubakar M. Water-borne diseases, cost of illness and willingness to pay for diseases interventions in rural communities of developing countries. Iran J Public Health. 2012;41(6):39-49. Epub 2012 Jun 30. PMID: 23113192; PMCID: PMC3469006.
- Ruggeri FM, Bonomo P, Ianiro G, Battistone A, Delogu R, Germinario C, Chironna M, Triassi M, Campagnuolo R, Cicala A, Giammanco GM, Castiglia P, Serra C, Gaggioli A, Fiore L. Rotavirus genotypes in sewage treatment plants and in children hospitalized with acute diarrhea in Italy in 2010 and 2011. Appl Environ Microbiol. 2015 Jan;81(1):241-9. doi: 10.1128/AEM.02695-14. Epub 2014 Oct 24. PMID: 25344240; PMCID: PMC4272723.
- World health statistics: monitoring health for the SDGs. Geneva: WHO. 2018.
- La Rosa G, Fratini M, della Libera S, Iaconelli M, Muscillo M. Emerging and potentially emerging viruses in water environments. Ann Ist Super Sanita. 2012;48(4):397-406. doi: 10.4415/ANN_12_04_07. PMID: 23247136.
- Allen WE, Altae-Tran H, Briggs J, Jin X, McGee G, Shi A, Raghavan R, Kamariza M, Nova N, Pereta A, Danford C, Kamel A, Gothe P, Milam E, Aurambault J, Primke T, Li W, Inkenbrandt J, Huynh T, Chen E, Lee C, Croatto M, Bentley H, Lu W, Murray R, Travassos M, Coull BA, Openshaw J, Greene CS, Shalem O, King G, Probasco R, Cheng DR, Silbermann B, Zhang F, Lin X. Population-scale longitudinal mapping of COVID-19 symptoms, behaviour and testing. Nat Hum Behav. 2020 Sep;4(9):972-982. doi: 10.1038/s41562-020-00944-2. Epub 2020 Aug 26. PMID: 32848231; PMCID: PMC7501153.
- Beetz C, Skrahina V, Förster TM, Gaber H, Paul JJ, Curado F, Rolfs A, Bauer P, Schäfer S, Weckesser V, Lieu V, Radefeldt M, Pöppel C, Krake S, Kandaswamy KK, Bruesehafer K, Vogel F.Rapid Large-Scale COVID-19 Testing during Shortages. Diagnostics. 2020;10(7). doi: 10.3390/diagnostics10070464.
- Boehm AB, Hughes B, Duong D, Chan-Herur V, Buchman A, Wolfe MK, White BJ. Wastewater concentrations of human influenza, metapneumovirus, parainfluenza, respiratory syncytial virus, rhinovirus, and seasonal coronavirus nucleic-acids during the COVID-19 pandemic: a surveillance study. Lancet Microbe. 2023 May;4(5):e340-e348. doi: 10.1016/S2666-5247(22)00386-X. Epub 2023 Mar 22. PMID: 36965504; PMCID: PMC10032662.
- Bidawid S, Farber JM, Sattar SA. Contamination of foods by food handlers: experiments on hepatitis A virus transfer to food and its interruption. Appl Environ Microbiol. 2000 Jul;66(7):2759-63. doi: 10.1128/AEM.66.7.2759-2763.2000. PMID: 10877765; PMCID: PMC92070.
- Forés E, Rusiñol M, Itarte M, Martínez-Puchol S, Calvo M, Bofill-Mas S. Evaluation of a virus concentration method based on ultrafiltration and wet foam elution for studying viruses from large-volume water samples. Sci Total Environ. 2022 Jul 10;829:154431. doi: 10.1016/j.scitotenv.2022.154431. Epub 2022 Mar 9. PMID: 35278558.
- Abd El-Daim SE, Mohamed Shaheen NF, Hosseney EN, Elhosainy AM, Nehal IA, Elmahdy ME, Ali MA. Molecular detection and genotyping of group a rotavirus by multiplex semi-nested rt-pcr in sewage water and sludge. Journal of Microbiology & Biotechnology. 2019;4(1):2576-7771. doi: 10.23880/oajmb-16000141.
- Hassine-Zaafrane M, Kaplon J, Ben Salem I, Sdiri-Loulizi K, Sakly N, Pothier P, Aouni M, Ambert-Balay K. Detection and genotyping of group A rotaviruses isolated from sewage samples in Monastir, Tunisia between April 2007 and April 2010. J Appl Microbiol. 2015 Nov;119(5):1443-53. doi: 10.1111/jam.12920. Epub 2015 Sep 24. PMID: 26248601.
- Dubois E, Le Guyader F, Haugarreau L, Kopecka H, Cormier M, Pommepuy M. Molecular epidemiological survey of rotaviruses in sewage by reverse transcriptase seminested PCR and restriction fragment length polymorphism assay. Appl Environ Microbiol. 1997 May;63(5):1794-800. doi: 10.1128/aem.63.5.1794-1800.1997. PMID: 9143113; PMCID: PMC168473.
- Shaheen MNF, Abd El-Daim SA, Ahmed NI, Elmahdy EM. Molecular detection of three gastroenteritis viruses in urban sewage treatment plant and river water in Egypt. Egypt. J. Aquat. Biol. Fish. 2018;22(5):615-627. doi: 10.21608/EJABF.2018.60278.
- Frank C, Walter J, Muehlen M, Jansen A, van Treeck U, Hauri AM, Zoellner I, Rakha M, Hoehne M, Hamouda O, Schreier E, Stark K. Major outbreak of hepatitis A associated with orange juice among tourists, Egypt, 2004. Emerg Infect Dis. 2007 Jan;13(1):156-8. doi: 10.3201/eid1301.060487. PMID: 17370535; PMCID: PMC2725821.
- MacDonald E, Steens A, Stene-Johansen K, Gillesberg Lassen S, Midgley S, Lawrence J, Crofts J, Ngui SL, Balogun K, Frank C, Faber M, Gertler M, Verhoef L, Koopmans M, Sane J, van Pelt W, Sundqvist L, Vold L. Increase in hepatitis A in tourists from Denmark, England, Germany, the Netherlands, Norway and Sweden returning from Egypt, November 2012 to March 2013. Euro Surveill. 2013 Apr 25;18(17):20468. Erratum in: Euro Surveill. 2013;18(18). pii: 20473. PMID: 23647624.
- Divizia M, Gabrieli R, Stefanoni ML, Renganathan E, El Ghazzawi E, Kader OA, Gamil F, El Sawaf G, El Sherbini E, Saleh E, Degener AM, Noce A, Zaratti L, Modesti A, Panà A. HAV and HEV infection in hospitalised hepatitis patients in Alexandria, Egypt. Eur J Epidemiol. 1999 Aug;15(7):603-9. doi: 10.1023/a:1007514030062. PMID: 10543349.
- Hamza H, Abd-Elshafy DN, Fayed SA, Bahgat MM, El-Esnawy NA, Abdel-Mobdy E. Detection and characterization of hepatitis A virus circulating in Egypt. Arch Virol. 2017 Jul;162(7):1921-1931. doi: 10.1007/s00705-017-3294-4. Epub 2017 Mar 16. PMID: 28303345.
- Miagostovich MP, Ferreira FF, Guimarães FR, Fumian TM, Diniz-Mendes L, Luz SL, Silva LA, Leite JP. Molecular detection and characterization of gastroenteritis viruses occurring naturally in the stream waters of Manaus, central Amazonia, Brazil. Appl Environ Microbiol. 2008 Jan;74(2):375-82. doi: 10.1128/AEM.00944-07. Epub 2007 Dec 7. PMID: 18065620; PMCID: PMC2223260.
- Villena C, El-Senousy WM, Abad FX, Pintó RM, Bosch A. Group A rotavirus in sewage samples from Barcelona and Cairo: emergence of unusual genotypes. Appl Environ Microbiol. 2003 Jul;69(7):3919-23. doi: 10.1128/AEM.69.7.3919-3923.2003. PMID: 12839761; PMCID: PMC165171.
- Rodríguez-Díaz J, Querales L, Caraballo L, Vizzi E, Liprandi F, Takiff H, Betancourt WQ. Detection and characterization of waterborne gastroenteritis viruses in urban sewage and sewage-polluted river waters in Caracas, Venezuela. Appl Environ Microbiol. 2009 Jan;75(2):387-94. doi: 10.1128/AEM.02045-08. Epub 2008 Nov 21. PMID: 19028907; PMCID: PMC2620703.
Content Alerts
SignUp to our
Content alerts.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.