Medicine Group . 2023 April 26;4(4):793-800. doi: 10.37871/jbres1737.
Development of Ratiometric Fluorescence Sensing Molecule for Caspase-14, a Key Enzyme of Epidermis Metabolism
Miho Suzuki*, Koki Taguma, Udari Kalpana Bandaranayake and Wataru Ikeda
Abstract
Skin is the vastest organ harboring principle functions to protect from external stimuli and internal moisture desorption. The loss of functions might bring about diseases and senescence and well maintained metabolisms are recognized as crucial points to prevent those situations. But due to complexities of skin tissues, there would be some difficulties in progress of elucidation for skin metabolisms in detail. Various differentiated tissues layered in order can form dermis and epidermis to exfoliate as scurf in the top layer finally. Caspase-14 categorized in cysteine-aspartic acid protease (Caspase) family that has been identified as key enzymes to promote programmed cell deaths by sequential activations is known to be specifically expressed in the upper tissues and involved in the differentiation, but not verified to have relationship with final exfoliation processes at the time of cell death. We then developed Fluorescence Resonance Energy Transfer (FRET) based sensing molecules to monitor Caspase-14 activity quantitatively. We utilized Green Fluorescent Protein (GFP) and organic fluorescent dyes because FRET phenomena could be achieved under condition with regulated interactions between at least two fluorescent molecules. The elaborately designed fluorescent complexes could render quantifiable sensing over fluorescent background but be simply introduced into cells via endocytic mechanisms to detect intracellular enzymatic activities alive. In this study, we demonstrated Caspase-14 activation during apoptosis induction in cultured normal epidermal cells that suggested possibilities of Caspase-14 contribution to programmed cell deaths.
Introduction
Skin is the largest body part for human to accomplish barrier functions to external stimuli such as UV, air pollutants and infectious living organisms etc and internal moisture loss. Body temperature control and maintenance of adequate electrolyte balance by perspiration are significant performances to keep healthy conditions as well. On the other hand, skin is complicated tissues consisting of hypodermis, dermis and epidermis including diverse differentiated stratums and regenerated by exfoliation of top layer, keratinocyte [1]. Though disorder of the skin metabolisms might cause diseases and senescence [2], difficulties in research development would happen as a result of complex skin organizations. The cysteine-aspartic acid protease (Caspase) family commonly commit to programed cell death progressions through sequential activation from the initiation to execution stages. Caspase-14 has already been known to be expressed in upper layer of epidermis and contribute to production of moisture factors [3] and enucleation for flattened by filaggrin digestion and densely packed keratinocyte [4] so that is considered to play significant roles in skin barriers formation [5], however, its commitment to programmed cell deaths including keratinocyte desquamation has not been proved [6]. In this study, we tried to generate Fluorescence Resonance Energy Transfer (FRET) based molecular sensor to detect and quantify activation of Caspase-14. The FRET based molecular sensor requiring at least two fluorescence constituents as donor and acceptor molecules could take advantages of overcome for fluctuated autofluorescence from cells to sense biological phenomena accurately using fluorescence materials [7,8]. In addition, the ratio of FRET efficiency exchanges be utilized to evaluate targets of sensing. So we have introduced Green Fluorescent Protein (GFP) as donor molecule and organic dyes as acceptor molecules. We have already exploited them to fabricate molecular sensors for Caspase-3 and Caspase-9 as dose or time dependent enzymatic profiles [9-11] and confirmed them applicable to similar molecules due to robust molecular design (Scheme 1). Moreover, our molecular sensors can be noninvasively introduced into cells via endocytic pathways [11]. We thus utilized epidermal cells, FEPE1L-8 with differentiation potential [12] to examine Caspase-14 activity during programmed cell death, apoptosis induction. Here we demonstrated that our developed molecular sensor for Caspase-14 activity could detect corresponding live activities during apoptosis progression in FEPE1L-8.
Materials and Methods
Plasmid construction for Caspase-14, Caspase-1 activity detection
Inverse Polymerase Chain Reaction (iPCR) [13] was used to construct plasmids containing the reporter genes for GFP mutants with Caspase-14 or Caspase-1 activity detections. Theses plasmid based on the pUV5cas12-1 [10] or pUV5casS52tag [9] plasmids were replaced its Caspase-9 sensitive sequences, TTAGAACATGAT with Caspase-14 recognition sequences, TGGGAACATGAT, or its Caspase-3 sensitive sequences, GATGAAGTAGAT with Caspase-1 recognition sequences, TATGTTGCAGAT respectively, where we altered amino acid sequences, LEHD for Caspase-9 or DEVD for Caspase-3 to Caspase-14 recognizable WEHD or Caspase-1 detectible YVAD [14], around C-terminus of their GFPs’ core. PCR performances were confirmed by sequencing, and the resulting plasmids were named pUV5cas-14 or pUV5cas-1 individually.
Fluorescent protein isolation
GFP variants were isolated and verified as previously described [9]. Briefly, obtained plasmids mentioned above (pUV5as-14, pUV5cas-1) were transformed into Escherichia coli BL-21(DE3) (Merck Biosciences, Darmstadt, Germany). E. coli from resulting single colony was grown at 37°C in Lysogeny Broth (LB) medium containing 75 μg/mL ampicillin for 12h. Subsequently, an aliquot of the culture medium was diluted 10 times with fresh LB medium supplemented with 75 μg/mL ampicillin, and cultured at 28°C for 48h while promoting protein expression with 1 mM of isopropyl β-D-thiogalactopyranoside (IPTG) (Takara Bio. Inc. Japan). The bacteria were harvested and lysed with bacterial protein extraction reagent B-PER II (Thermo Fisher Scientific Inc., Waltham, MA, USA), according to the manufacturer’s instructions. The lysates were centrifuged at 15,000 rpm for 15 min at room temperature. The supernatant was applied to Ni2+-NTA resin (Qiagen, Düsseldorf, Germany), and the His-tagged fluorescent proteins were isolated using affinity chromatography according to standard procedures for purification of histidine tag-fused proteins. The recovered protein was reconstituted in Phosphate-Buffered Saline (PBS) and subjected to Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis (SDS-PAGE) to determine purity and oligomerization. The purified protein was further quantified and qualified by spectrophotometry (DU 640 Beckman Colter Inc. Japan), with absorbance at 280 nm and 488 nm.
Sensing molecule preparation
The sensing molecule was conducted as previously described [15]. Briefly, the purified proteins were chemically modified with fluorescent dyes, Alexa Fluor 546 (Life Technologies Corp., Carlsbad, CA, USA). The protein was dissolved in PBS (100 μL) containing 1 mM Dithiothreitol (DTT) to give a final concentration of 10 μM, which was then incubated for 10 min at room temperature. DTT was removed by gel filtration (NICK column, GE Healthcare UK Ltd., Chalfont St Giles, UK). An aliquot of the eluate (400 μL) was immediately mixed with 0.8 μL fluorescent dye in Dimethyl sulfoxide (DMSO) solution (10 mg/mL), and the mixture was incubated at 37°C for 4h following further incubation at 4°C over night. Excess dye was removed by ultrafiltration using Amicon Ultra-0.5 10 K (Merck KGaA, Darmstadt, Germany). The fluorescence spectrum was measured at an excitation wavelength of 488 nm using a Shimadzu RF-5300PC Spectro fluorophotometer (Shimadzu, Kyoto, Japan). We have already demonstrated FRET efficiencies of our corresponding sensing molecules could be approximated by the ratio of fluorescence intensity maxima for donor and acceptor due to high FRET efficiency, though accurate FRET efficiency is defined as the probability of donor molecules transferring fluorescent excitation energy to acceptor molecules. Moreover, approximate FRET efficiencies can be applied to normalize Caspase-14 or Caspase-1 activities.
Determining Caspase‑14 activity and specificity assays with sensing molecule in vitro
We determined Caspase-14 sensing in vitro according to a previously established approach [15] with the Alexa Fluor 546-UVcas14 conjugate sensing molecule or Alexa Fluor 546-UV5cas-1 conjugate sensing molecule. Here, 0.2 units of Caspase-14 (Thermo Fisher Scientific Inc., Waltham, MA, USA) were added to 20 μL of 5.0 μM sensing molecule solution reconstituted in an assay buffer containing 100 mM 2-[4-(2-hydroxyethyl)piperazin-1- yl]-ethane sulfonic acid (HEPES), 60 mM NaCl, 0.01% 3-[(3- cholamidopropyl)dimethylammonio]-1-propanesulfonate(CHAPS) detergent, 5 mM DTT, 1% glycerol, and 1.1 M sodium citrate [16]. We prepared the corresponding sample without Caspase-14. Both solutions were then incubated at 37°C for 2 h and checked emission patterns upon 20 times dilution with PBS using fluorescent spectroscopy (Shimadzu RF-5300PC, Shimadzu, Kyoto, Japan). We estimated ratio of FRET efficiency exchanges from FRET efficiencies between sample with or without Caspase-14 treatment.
Determining Caspase‑1 activity and specificity assays with sensing molecule in vitro
We validated Caspase-1 sensing in vitro in the same way as Caspase-14 mentioned above using prepared Alexa Fluor 546- UV5cas-1 conjugate sensing molecule and specific buffer for Caspase-1 instead. In short, 1.0 units of Caspase-1 (Merck Biosciences, Darmstadt, Germany) were added to 20 μL of 2.0 μM sensing molecule solution reconstituted in an assay buffer containing 50mM 2-[4-(2-hydroxyethyl)piperazin-1- yl]-ethane sulfonic acid (HEPES), 100 mM NaCl, 0.1% 3-[(3- cholamidopropyl)dimethylammonio]-1-propanesulfonate(CHAPS) detergent, 10mM DTT, 10% glycerol, and 1 mM ethylenediaminetetraacetic acid [17]. We prepared the corresponding sample without Caspase-1. Both solutions were then incubated at 37°C for 2h and checked emission patterns upon 20 times dilution with PBS using fluorescent spectroscopy (Shimadzu RF-5300PC, Shimadzu, Kyoto, Japan). We estimated ratio of FRET efficiency exchanges from FRET efficiencies between sample with or without Caspase-1 treatment.
Sensing molecule introduction into epidermal cells with differentiation ability
FEPE1L-8, normal epidermal cells with differentiation ability were cultured with HuMedia KB-2 (Kurabo Industries Ltd. Kurashiki, Japan) supplemented with 10 ug/mL of insulin, 0.1 ng/mL of hEGF, 0.67 ug/mL of Hydrocortisone hemisuccinate, 50 ug/mL of gentamicin, 50 ng/mL of amphotericin B, 0.4% V/V of Bovine Pituitary Gland Extract (BPE) [12]. Confluent cells were washed 3 times with fresh culture medium and added with aliquots of which containing 5 μM corresponding sensing molecules for 12h incubation at 37°C under an atmosphere containing 5% CO2. The residual sensing molecules were removed by washing the cells with fresh culture medium.
Apoptosis induction for epidermal cells with differentiation ability
Induction of apoptosis in FEPE1L-8 was conducted equally for other cell types [15]. Briefly, culture medium for sensing molecule uptake FEPE1L-8 cells was replaced with fresh culture medium containing 100 ng/mL and 1.0 mg/mL of tumor necrosis factor alpha (TNF-α) and 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione (cycloheximide) were respectively and incubated for 12h incubation at 37°C under an atmosphere containing 5% CO2.
Flow cytometry analysis
After sensing molecule introduction or apoptosis induction, cells were immediately collected and analyzed using flow cytometry. The culture supernatant with sensing molecules or apoptosis inducing reagents was removed and rinsed with 150 μL of 0.2 mM EDTA. The chelating solution was stored and incubated with 150 μL each of Accutase and Accumax cell detachment mixtures (Innovative Cell technologies, San Diego, CA, USA) for 10 min at 37°C. The detached mixtures with the detached cells were stored as recovered cell solutions to be combined with stored chelating solutions. Trypsin (0.25%, 250 μL) was then added to the culture dish for complete cell detachment. The resulting solutions with cell debris were joined with those previously stored and the whole solutions were subjected to flow cytometry analysis using a SONY SH800Z (SONY Corp., Tokyo, Japan). The data obtained were processed by FLOWJO software (Becton, Dickinson and Company, San Jose, CA, USA).
Results and Discussion
Caspase-14 activity detection with sensing molecule in vitro
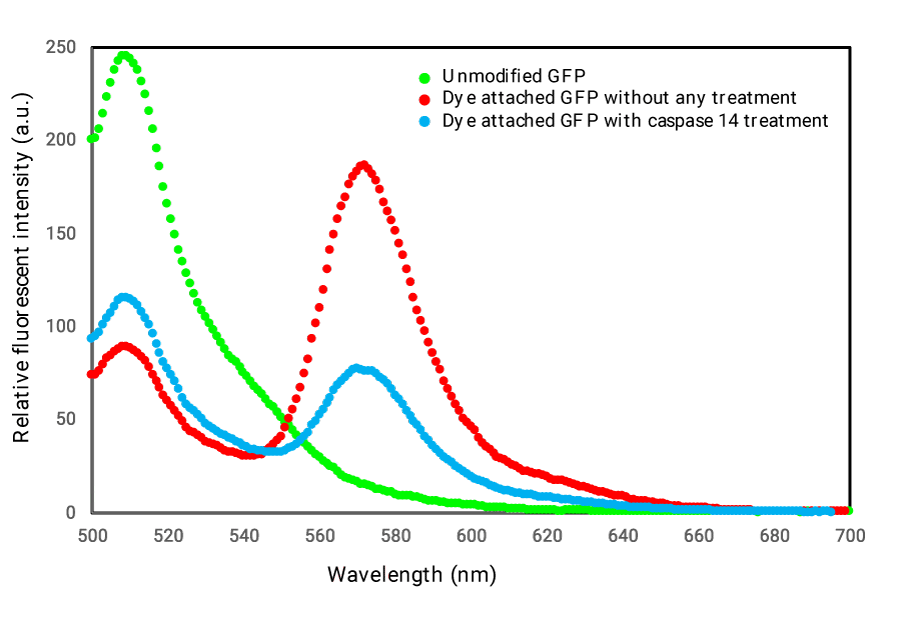
After preparation of sensing molecule for Caspase-14 activity, we investigated the performance through emission profiles of obtained GFP-Alexa dye complexes (Figure 1). We also demonstrated representative emission pattern of unmodified GFP as reference. Fluorescent profile for the modified GFP as shown in the figure 1 was drastically changed. We could estimate FRET efficiencies as describe in the experimental section. During three times trial of preparations for the sensing molecule, we obtained average FRET efficiencies calculated as 1.88±0.73. The value might be high enough for live monitoring cellular events in side cells. We then checked sensing properties upon Caspase-14 treatment (Figure 1). As shown in figure 1, sensing molecule treated with Caspase-14 indicated alteration in its emission pattern to corresponding pattern without Caspase-14 handling as FRET disappearance. We approximated FRET efficiency exchanges for the emission profiles with or without Caspase-14 from 3 times equivalent experiments as well to get 2.07 ± 0.97. In consideration of performances of related sensing molecules for other Caspase activities [9-11], the FRET efficiency exchanges we got for Caspase-14 might meet the criteria of sensitivities to detect biological phenomena inside cell.
Introduction of sensing molecule for Caspase-14 activity into normal epidermal cells with differentiation, FEPE1L-8
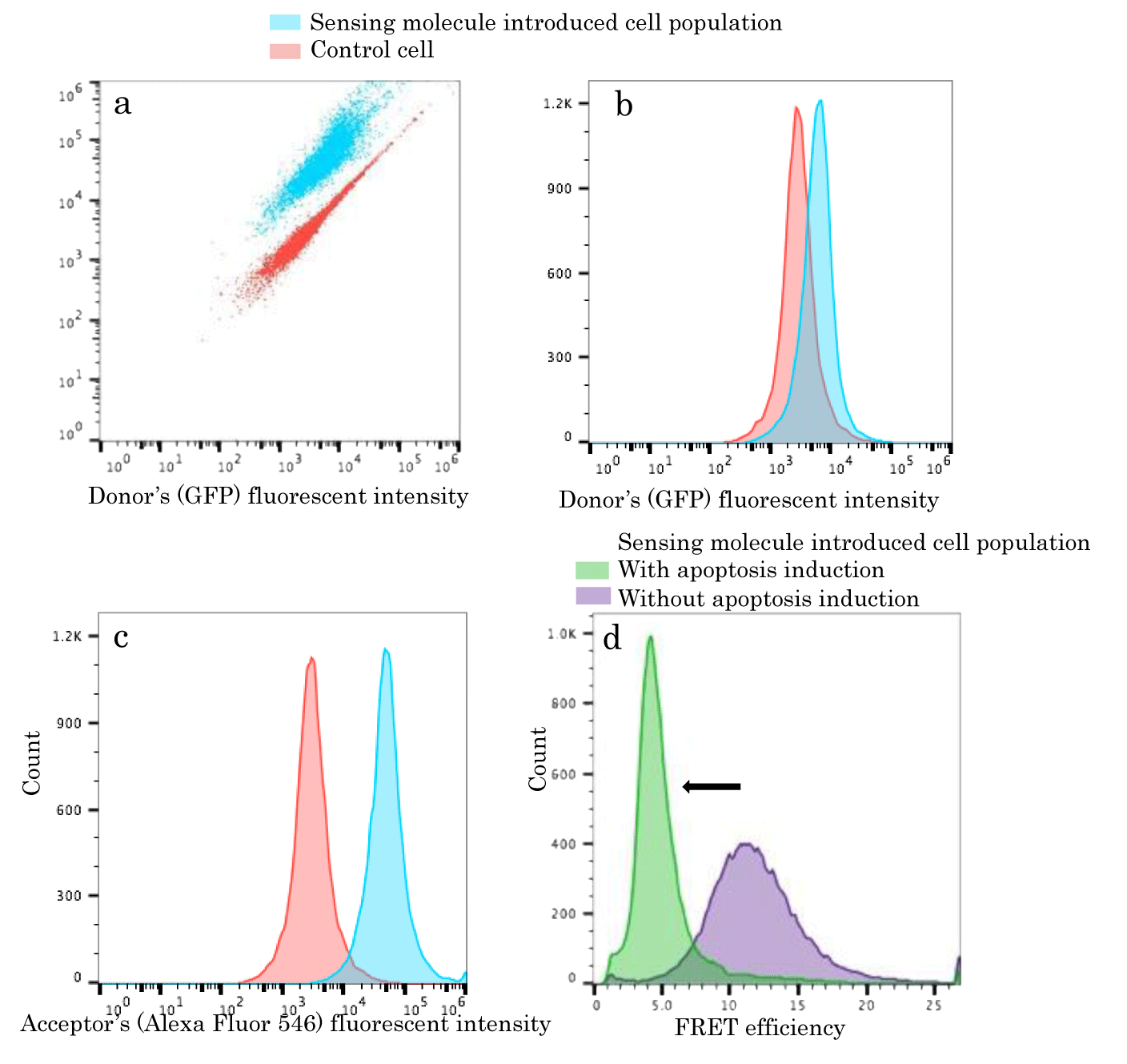
As we verified sufficient behavior of our sensing molecule for Caspase-14 activity in vitro, we attempted to introduce the molecule into normal epidermal cells, FEPE1L-8 simply. We just mixed appropriate amount of sensing molecule with cultured medium of the cells for 12h denoted in the experimental section. We next harvested resulting cells to be analyzed with flow cytometry. Reference cell populations without sensing molecule uptake were equally harvested and applied to flow cytometric analysis. As shown in dot plots of figure 2a, we could acquire two distinctive cell populations. The reference cell populations were mainly distributed left lower area compared to sensing molecule introduced cell populations. Relative fluorescence intensities in both donor and acceptor component increased for cell populations with sensing molecules. Donor and acceptor fluorescent components were separately evaluated as histograms (Figures 2b,c). Due to high FRET efficiency of sensing molecules, acceptor fluorescent component intensities derived from sensing molecule uptake cell populations were much higher than reference ones. It was indicated that our GFP-Alexa dye complex was efficiently introduced into cells as it was. This suggested the sensing molecule might be suitable to monitor time dependent cellular events alive as represented in the preceding reports [9-11].
Caspase-14 activity monitoring upon apoptosis induction to FEPE1L-8
As we determined occurrences of sensing molecule enough amount applicable to target detection inside cells, we induced programmed cell deaths, apoptosis to those cells. We prepared cells with sensing molecule uptakes but not apoptotic induction as well. We then estimated average FRET efficiencies for each cell populations shown in histograms of figure 2d. We observed so clear distribution shift to FRET efficiencies cancellation upon apoptosis inductions that sensing molecules inside apoptotic cells could detect Caspase-14 activations. It was suggested programmed cell death induced normal epidermal cells might also activate Caspase-14 contributed to other cellular events for proper skin maintenances. Those direct indications for probable contributions of Caspase-14 to programmed cell death could hardly be done.
We then examined properties of our sensing molecules more on the point of specificity for validation of our findings.
Caspase-1 activity detection with sensing molecule in vitro
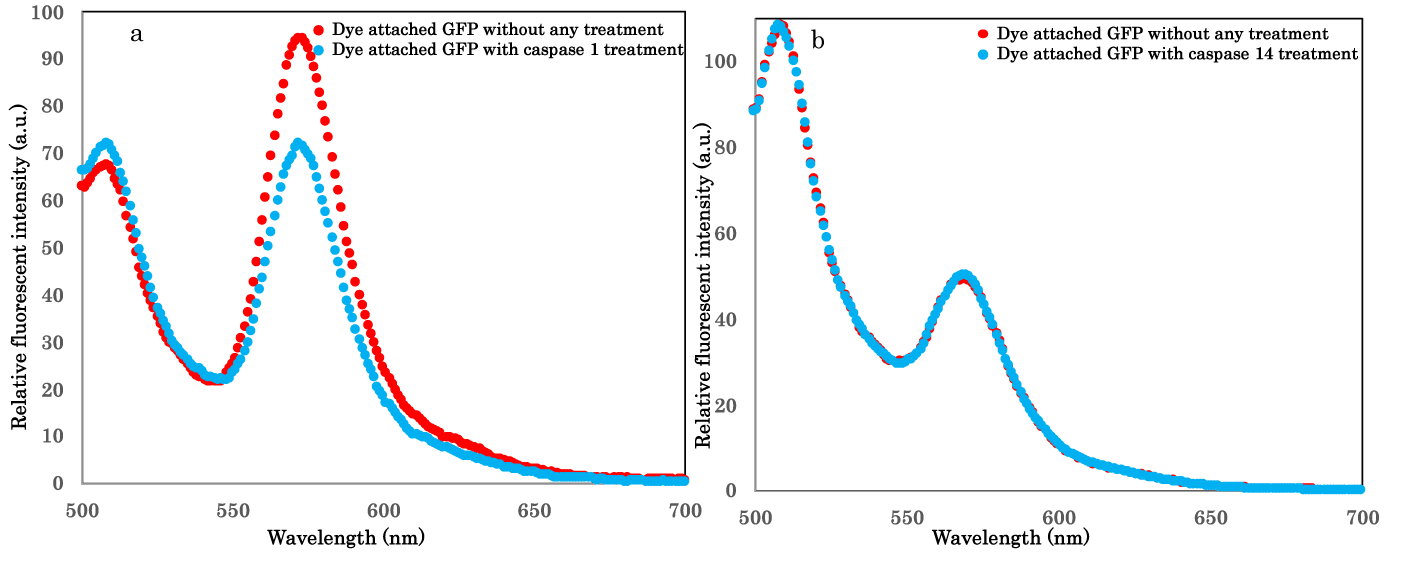
Substrates and inhibitors for most Caspases were reviewed for cell deaths study [14] based on many related studies. Adopted sequence in this study for sensing molecule, WEHD, was regarded as good substrate for Caspase-14 but common substrate for Caspase-1 [17]. In contrast, YVAD was presumed as poor for Caspase-14 but good for Caspase-1 activity detections [18]. Thus, we checked additional sensing molecule including YVAD to detect specific Caspase-1 activity in vitro for aimed next research on epidermis metabolism in terms of cell deaths. We investigated the sensing molecule not to detect Caspase-14 activity either. As shown in figures 3a,b, resulting sensing molecules could detect Caspase-1 activity but not Caspase-14 one in vitro. It was demonstrated that our sensing molecules could consistently functions as reported peptide based or other constructed substrates. However, easy accesses for live cell imaging of our sensing molecules would be advantageous for cellular dynamic studies.
Conclusion
We have developed FRET based sensing molecule consisting of green fluorescent protein and fluorescent organic dye applied to live activity detection specific for key protease classified in cysteine-aspartic acid protease (Caspase) family, Caspase-14 inside cells. Though Caspase-14’s performances were verified as commitment to epidermis barrier functions, it has not been found out to be involved in desquamation as programmed cell deaths during normal skin cycle of keratinocyte skin cells being born from stem cells at the basal layer of the epidermis, proliferation, differentiation and detachment from the skin's surface [5,6]. In this study, we could identify possibility for participation of Caspase-14 in programmed cell deaths at epidermis. Proteomic study rendered capase-14 expression profiles during epidermis metabolism, however, monitoring live activation of Caspase-14 upon programmed cell death induction using normal keratinocyte might be significant. Considering epidermis functions, relationship between Caspase-14 activation and other cell deaths stimulations as exposures to ROS, UV and air pollutant that might cause other types of programmed cell deaths, as pyroptosis, necroptois, netosis, ferroptosis etc should be examined as next step to ascertain engrossment to skin exfoliation. From these points, our success to establish additional sensing molecule for specific Caspase-1 activity detection can promote discriminations of cell death types occurred upon diverse stimulations to epidermis. Development of another corresponding sensing molecule for Caspase-14 activity could be more specific and practical to determine functions of Caspase-14 inside epidermis.
Acknowledgment
This study was kindly supported by Nippi Incorporated including FEPE1L-8 cells offer. Moreover, we gratefully acknowledge the work of past and present members of our laboratory.
Conflicts of Interest
The authors have no conflicts of interest to declare.
References
- Hofmann E, Schwarz A, Fink J, Kamolz LP, Kotzbeck P. Modelling the Complexity of Human Skin in vitro. Biomedicines. 2023 Mar 6;11(3):794. doi: 10.3390/biomedicines11030794. PMID: 36979772; PMCID: PMC10045055.
- Wang J, Cui B, Chen Z, Ding X. The regulation of skin homeostasis, repair and the pathogenesis of skin diseases by spatiotemporal activation of epidermal mTOR signaling. Front Cell Dev Biol. 2022 Jul 22;10:950973. doi: 10.3389/fcell.2022.950973. PMID: 35938153; PMCID: PMC9355246.
- Hoober JK, Eggink LL. The Discovery and Function of Filaggrin. Int J Mol Sci. 2022 Jan 27;23(3):1455. doi: 10.3390/ijms23031455. PMID: 35163390; PMCID: PMC8835998.
- Quiroz FG, Fiore VF, Levorse J, Polak L, Wong E, Pasolli HA, Fuchs E. Liquid-liquid phase separation drives skin barrier formation. Science. 2020 Mar 13;367(6483):eaax9554. doi: 10.1126/science.aax9554. PMID: 32165560; PMCID: PMC7258523.
- Markiewicz A, Sigorski D, Markiewicz M, Owczarczyk-Saczonek A, Placek W. Caspase-14-From Biomolecular Basics to Clinical Approach. A Review of Available Data. Int J Mol Sci. 2021 May 25;22(11):5575. doi: 10.3390/ijms22115575. PMID: 34070382; PMCID: PMC8197544.
- Anderton H, Alqudah S. Cell death in skin function, inflammation, and disease. Biochem J. 2022 Aug 12;479(15):1621-1651. doi: 10.1042/BCJ20210606. PMID: 35929827; PMCID: PMC9444075.
- Fang C, Huang Y, Zhao Y. Review of FRET biosensing and its application in biomolecular detection. Am J Transl Res. 2023 Feb 15;15(2):694-709. PMID: 36915763; PMCID: PMC10006758.
- Wu Y, Jiang T. Developments in FRET- and BRET-Based Biosensors. Micromachines (Basel). 2022 Oct 20;13(10):1789. doi: 10.3390/mi13101789. PMID: 36296141; PMCID: PMC9610962.
- Suzuki M, Tanaka S, Ito Y, Inoue M, Sakai T, Nishigaki K. Simple and tunable Förster resonance energy transfer-based bioprobes for high-throughput monitoring of caspase-3 activation in living cells by using flow cytometry. Biochim Biophys Acta. 2012 Feb;1823(2):215-26. doi: 10.1016/j.bbamcr.2011.07.006. Epub 2011 Jul 21. PMID: 21791227.
- Suzuki M, Sakata I, Sakai T, Tomioka H, Nishigaki K, Tramier M, Coppey-Moisan M. A high-throughput direct fluorescence resonance energy transfer-based assay for analyzing apoptotic proteases using flow cytometry and fluorescence lifetime measurements. Anal Biochem. 2015 Dec 15;491:10-7. doi: 10.1016/j.ab.2015.08.022. Epub 2015 Sep 1. PMID: 26334608.
- Suzuki M, Shindo Y, Yamanaka R, Oka K. Live imaging of apoptotic signaling flow using tunable combinatorial FRET-based bioprobes for cell population analysis of caspase cascades. Sci Rep. 2022 Dec 7;12(1):21160. doi: 10.1038/s41598-022-25286-z. PMID: 36476686; PMCID: PMC9729311.
- Carter WG, Ryan MC, Gahr PJ. Epiligrin, a new cell adhesion ligand for integrin alpha 3 beta 1 in epithelial basement membranes. Cell. 1991 May 17;65(4):599-610. doi: 10.1016/0092-8674(91)90092-d. PMID: 2032285.
- Imai Y, Matsushima Y, Sugimura T, Terada M. A simple and rapid method for generating a deletion by PCR. Nucleic Acids Res. 1991 May 25;19(10):2785. doi: 10.1093/nar/19.10.2785. PMID: 1645866; PMCID: PMC328208.
- Julien O, Wells JA. Caspases and their substrates. Cell Death Differ. 2017 Aug;24(8):1380-1389. doi: 10.1038/cdd.2017.44. Epub 2017 May 12. PMID: 28498362; PMCID: PMC5520456.
- Suzuki M, Ito Y, Sakata I, Sakai T, Husimi Y, Douglas KT. Caspase-3 sensitive signaling in vivo in apoptotic HeLa cells by chemically engineered intramolecular fluorescence resonance energy transfer mutants of green fluorescent protein. Biochem Biophys Res Commun. 2005 May 6;330(2):454-60. doi: 10.1016/j.bbrc.2005.02.178. PMID: 15796904.
- Mikolajczyk J, Scott FL, Krajewski S, Sutherlin DP, Salvesen GS. Activation and substrate specificity of caspase-14. Biochemistry. 2004 Aug 17;43(32):10560-9. doi: 10.1021/bi0498048. PMID: 15301553.
- Garcia-Calvo M, Peterson EP, Leiting B, Ruel R, Nicholson DW, Thornberry NA. Inhibition of human caspases by peptide-based and macromolecular inhibitors. J Biol Chem. 1998 Dec 4;273(49):32608-13. doi: 10.1074/jbc.273.49.32608. PMID: 9829999.
- Hibino T, Fujita E, Tsuji Y, Nakanishi J, Iwaki H, Katagiri C, Momoi T. Purification and characterization of active caspase-14 from human epidermis and development of the cleavage site-directed antibody. J Cell Biochem. 2010 Feb 15;109(3):487-97. doi: 10.1002/jcb.22425. PMID: 19960512.
Content Alerts
SignUp to our
Content alerts.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.