Medicine Group . 2023 April 27;4(4):801-805. doi: 10.37871/jbres1738.
Heads or Tails: Incomplete Penetrance and Variable Expressivity at 15q11.2 Microdeletion (BP1-BP2)
Teresa Enrique-Benedito1*, Vanesa Senoret1, Ana Nevot-Flor2, Natalia Artunedo-Jimenez1 and M Arrizabalaga1
2Neuropediatrics Department, Hospital Universitario de la Plana, Spain
- 15q11.2 microdeletion
- Array CGH
- Developmental delay
- Incomplete penetrance
- Variable expressivity
Abstract
Neurodevelopmental disorders are a group of pathologies with increased frequency in pediatric practice. During the last decade, genetic studies such as CGH arrays have aided in the search for genetic gains or losses that may help explain the symptoms.
There are CNVs, like the 15q11.2 microdeletion between BP1 and BP2, that although considered pathological, may present incomplete penetrance and variable expression, both of which add difficulty to the already complicated task of genetic counseling to the family members.
Hereby we present cases of three patients with similar phenotypes in which microdeletion of 15q11.2 was detected.
Introduction
Developmental delay, psychological issues such as Attention Deficit and Hyperactivity Disorder (ADHD), Autism Spectrum Disorder (ASD) and obsessive-compulsive behavior, intellectual disabilities, epilepsy and congenital abnormalities are some of disorders tested using the Array CGH technique [1].
The implementation of Array CGH in human genetic diagnosis has allowed the detection of submicroscopic Copy Number Variation (CNVs) associated with neurodevelopmental disorders. Some of these detected CNVs are classified as pathogenic or likely pathogenic, and for those where the relationship with the phenotype is not clear, they are known as Variants of Unknown Significance (VOUS) [2].
Previous publications have reported patients with 15q11.2 microdeletion between BP1 and BP2 breakpoints presenting developmental delay, intellectual disability, speech impairment, learning disabilities and/or behavioral disturbances [3]. This copy variant is known as chromosomic 15q11.2 deletion syndrome (#OMIM 615656) with typically encompasses a 300-500 Kb deletion, involving four not imprinted highly conserved genes; TUBGCP5, NIPA1, NIPA2 and CYFIP1. The latter three are widely expressed in the central nervous system and are associated with several brain disorders [4,5]. It is difficult to establish the clinical significance of this microdeletion due to incomplete penetrance since some carriers are healthy and variable expressivity found in mildly affected carriers [6].
It has been noticed that duplications of the BP1-BP2 can cause similar symptoms although deletions can have a greater impact [7].
Here we present three new cases in which the 15q11.2 deletion is present with above mentioned clinical symptomatology.
Materials and Methods
Patients
A total of 887 patients were referred for array CGH from March 2019 to October 2022 due to congenital abnormalities, behavioral issues, language and developmental delay in Centro Diagnostico Calderon. A report was obtained from each patient detailing symptoms and phenotype.
Array Comparative Genomic Hybridization (Array CGH)
Genomic DNA was extracted from the patients’ peripheral blood using qiaamp DNA blood mini kit (Qiagen). DNA concentration and quality was determined with NanoDrop spectrophotometer (Thermo Fisher Scientific). Detection of copy number variation was performed by comparative genomic hybridization (Array CGH) on a Cytoarray Plus (180K) microarray based on the construction of the NCBI37 (hg19) genome sequence using the Agilent Technologies platform. The labeling and hybridization of the sample has been carried out together with a diploid commercial reference DNA without alterations. The microarray used contains more than 180,000 probes, specially designed for the detection of copy number alterations in regions related to intellectual disability and/or developmental delay, polymalformation syndromes and autism, among others. The resolution limit of the array is ≥0.002Mb. The array design is based on ISCA v2.
The analysis of the results was carried out using the Agilent CytoGenomics 5.1.2.1 software and the ADM-2 alteration detection algorithm with a threshold value of 6.0. Those presenting a minimum of five consecutive probes are accepted as alterations.
Results
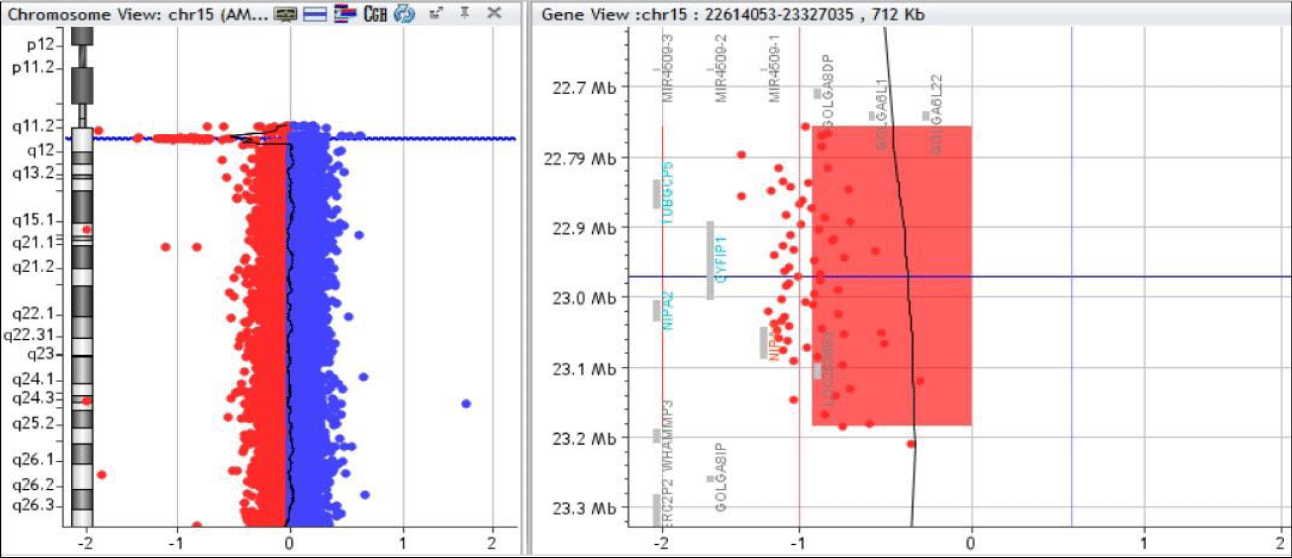
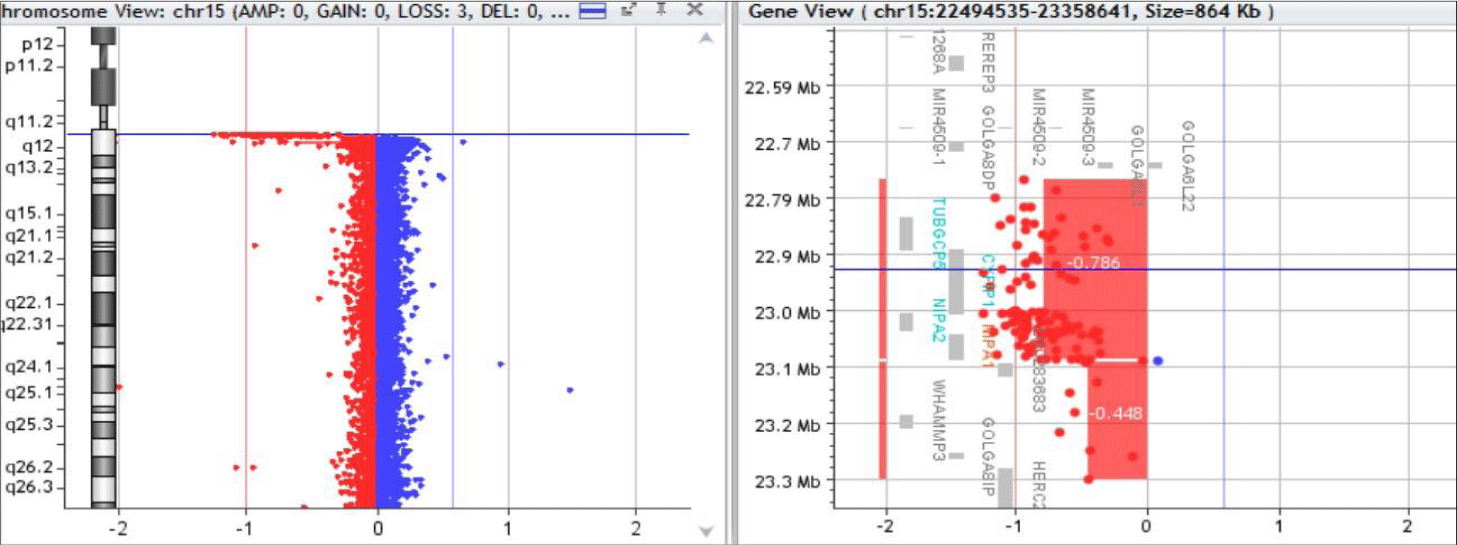
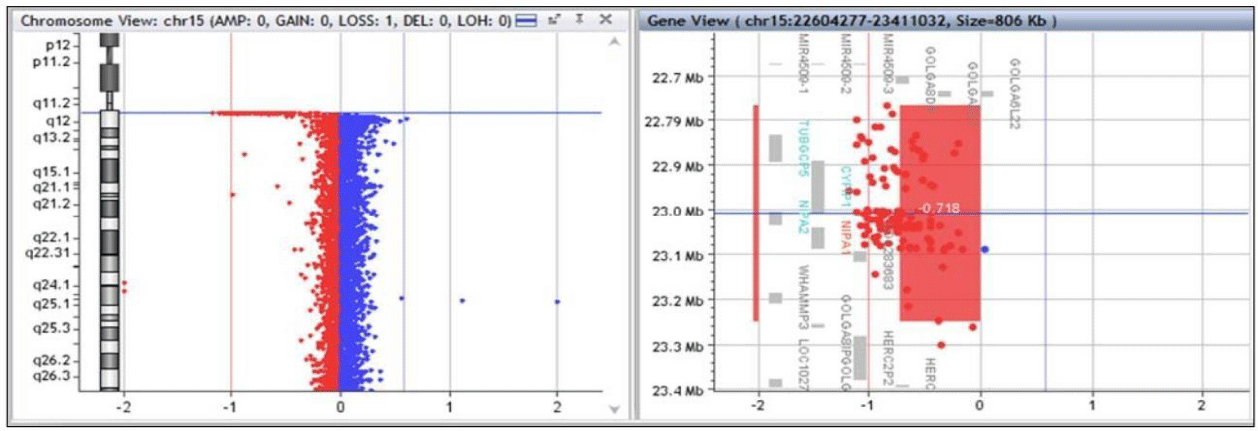
From 887 patients, we identified a 15q11.2 deletion between BP1-BP2 in 3 patients. The deletion involved the same four genes in all cases, ranging from 0.322 Mb to 0.484 Mb.
Two of the cases had parental origin (case 1 and 2), in case 3 the inheritance could not be verified.
Case 1 was a 12-year-old boy, who was under follow-up for malnutrition, borderline intellectual capacity, oppositional defiant disorder, and attention deficit hyperactivity disorder. He was prescribed drugs and there was an improvement in his behavior at school, but in the absence of it he was more upset. Previous karyotype and X-fragile studies were done, these being negative. Parents had no abnormal phenotype.
Array CGH 180k detected a 15q11.2 microdeletion of 0.428 Mb (chr15:22756650_23184439) (Figure 1). Additionally, a 7q31.1 deletion of unknown meaning was found together with other benign or probably benign duplications and deletions. His mother had the same 15q11.2 and 7q31.1 deletions.
Case 2 was a 11-year-old girl, who was being followed up for short stature, mild intellectual disability with mixed emotional and behavioral disturbances, and oppositional defiant disorder. She had speech impairment, insecurity and low self-esteem and suffered from obesity, with a weight of 52.6 kg and height of 128 cm. Moreover, she had peculiar phenotype with wide nasal bridge. Parents had no abnormal phenotype.
Array CGH 180k detected a 15q11.2 microdeletion of 0.322Mb (chr:22765628_23087554) was inherited from the father (Figure 2). A deletion at 20p12.2 of unknown significance was inherited from the mother. Other benign or probably benign duplications and deletions were found.
Case 3 was a 13-year-old girl, who was derived for difficulties in the development of language and oral comprehension. She had cognitive and attention disturbances.
She suffered from obesity with a weight of 99.5 kg and a height of 151.2 cm. She had karyotype study performed and this was found normal.
Array CGH 180k detected a 15q11.2 microdeletion of 0.484 Mb (chr15:22765628_23249681). Additionally, a deletion in 22q11.22 of unknown meaning was found together with other benign or probably benign duplications and deletions (Figure 3). The mother had cognitive impairment. However, heritability of the found deletions could not be tested.
Discussion
In this study we report 3 new cases with a 15q11.2 microdeletion between BP1 and BP2 of the Prader-Willi/Angelman critical region. Our patients had a microdeletion ranging from 322 kb to 484 kb involving four genes: TUBGCP5, NIPA1, NIP2 and CYF1P1.
All patients with the 15q11.2 deletion showed mild intellectual disabilities, speech impairment, and behavioural problems such as attention deficit and hyperactivity disorder between others.
The 15q11.2 microdeletion had been described in patients with moderate delayed psychomotor development, speech impairment, attention deficit and hyperactivity disorder, autism spectrum disorder and obsessive-compulsive disorder [3,8].
NIPA1, NIPA2 and CYF1P1 are widely expressed in the nervous system. Therefore, they play a role in neurodevelopment and have been associated with several brain disorders [2]. However, the haploinsufficiency of these genes alone does not explain the variable expressivity and incomplete penetrance observed in patients with mild phenotype [4], and in healthy carriers of the microdeletion [9]. An attempt has been made to explain this anomalous penetrance. One hypothesis named “two-hit model” argues that a second event could occur causing the haploinsufficiency of the remaining allele. Otherwise, a mutation in a modifier gene could affect the expression of these genes. Moreover, epigenetic modification and environmental implication should not be dismissed [3].
Conclusion
In conclusion, we present 3 new cases with 15q11.2 deletion with mild intellectual disability, oppositional defiant disorder, speech impairment and attention deficit hyperactivity disorder. Numerous symptoms are associated with this deletion in affected patients with healthy parental carriers. This supports variable expressivity and incomplete penetrance previously reported. Based on current ACMG guidelines this CNV would be consider pathogenic. Nevertheless, due to variable expressivity and incomplete penetrance this microdeletion could be considered as a susceptibility locus for neuropsychiatric and neurodevelopmental disorders. This fact turns genetic counseling into a "heads or tails" thus complicating genetic counseling, especially in prenatal cases, where the carrier status would not help to predict the phenotype of the future child.
References
- Gene expression microarray-Bioarray.
- Writing Committee for the ENIGMA-CNV Working Group; van der Meer D, Sønderby IE, Kaufmann T, Walters GB, Abdellaoui A, Ames D, Amunts K, Andersson M, Armstrong NJ, Bernard M, Blackburn NB, Blangero J, Boomsma DI, Brodaty H, Brouwer RM, Bülow R, Cahn W, Calhoun VD, Caspers S, Cavalleri GL, Ching CRK, Cichon S, Ciufolini S, Corvin A, Crespo-Facorro B, Curran JE, Dalvie S, Dazzan P, de Geus EJC, de Zubicaray GI, de Zwarte SMC, Delanty N, den Braber A, Desrivieres S, Di Forti M, Doherty JL, Donohoe G, Ehrlich S, Eising E, Espeseth T, Fisher SE, Fladby T, Frei O, Frouin V, Fukunaga M, Gareau T, Glahn DC, Grabe HJ, Groenewold NA, Gústafsson Ó, Haavik J, Haberg AK, Hashimoto R, Hehir-Kwa JY, Hibar DP, Hillegers MHJ, Hoffmann P, Holleran L, Hottenga JJ, Hulshoff Pol HE, Ikeda M, Jacquemont S, Jahanshad N, Jockwitz C, Johansson S, Jönsson EG, Kikuchi M, Knowles EEM, Kwok JB, Le Hellard S, Linden DEJ, Liu J, Lundervold A, Lundervold AJ, Martin NG, Mather KA, Mathias SR, McMahon KL, McRae AF, Medland SE, Moberget T, Moreau C, Morris DW, Mühleisen TW, Murray RM, Nordvik JE, Nyberg L, Olde Loohuis LM, Ophoff RA, Owen MJ, Paus T, Pausova Z, Peralta JM, Pike B, Prieto C, Quinlan EB, Reinbold CS, Reis Marques T, Rucker JJH, Sachdev PS, Sando SB, Schofield PR, Schork AJ, Schumann G, Shin J, Shumskaya E, Silva AI, Sisodiya SM, Steen VM, Stein DJ, Strike LT, Tamnes CK, Teumer A, Thalamuthu A, Tordesillas-Gutiérrez D, Uhlmann A, Úlfarsson MÖ, van 't Ent D, van den Bree MBM, Vassos E, Wen W, Wittfeld K, Wright MJ, Zayats T, Dale AM, Djurovic S, Agartz I, Westlye LT, Stefánsson H, Stefánsson K, Thompson PM, Andreassen OA. Association of Copy Number Variation of the 15q11.2 BP1-BP2 Region With Cortical and Subcortical Morphology and Cognition. JAMA Psychiatry. 2020 Apr 1;77(4):420-430. doi: 10.1001/jamapsychiatry.2019.3779. PMID: 31665216; PMCID: PMC6822096.
- Vanlerberghe C, Petit F, Malan V, Vincent-Delorme C, Bouquillon S, Boute O, Holder-Espinasse M, Delobel B, Duban B, Vallee L, Cuisset JM, Lemaitre MP, Vantyghem MC, Pigeyre M, Lanco-Dosen S, Plessis G, Gerard M, Decamp M, Mathieu M, Morin G, Jedraszak G, Bilan F, Gilbert-Dussardier B, Fauvert D, Roume J, Cormier-Daire V, Caumes R, Puechberty J, Genevieve D, Sarda P, Pinson L, Blanchet P, Lemeur N, Sheth F, Manouvrier-Hanu S, Andrieux J. 15q11.2 microdeletion (BP1-BP2) and developmental delay, behaviour issues, epilepsy and congenital heart disease: a series of 52 patients. Eur J Med Genet. 2015 Mar;58(3):140-7. doi: 10.1016/j.ejmg.2015.01.002. Epub 2015 Jan 14. PMID: 25596525.
- Doornbos M, Sikkema-Raddatz B, Ruijvenkamp CA, Dijkhuizen T, Bijlsma EK, Gijsbers AC, Hilhorst-Hofstee Y, Hordijk R, Verbruggen KT, Kerstjens-Frederikse WS, van Essen T, Kok K, van Silfhout AT, Breuning M, van Ravenswaaij-Arts CM. Nine patients with a microdeletion 15q11.2 between breakpoints 1 and 2 of the Prader-Willi critical region, possibly associated with behavioural disturbances. Eur J Med Genet. 2009 Mar-Jun;52(2-3):108-15. doi: 10.1016/j.ejmg.2009.03.010. Epub 2009 Mar 27. PMID: 19328872.
- Madrigal I, Rodríguez-Revenga L, Xunclà M, Milà M. 15q11.2 microdeletion and FMR1 premutation in a family with intellectual disabilities and autism. Gene. 2012 Oct 15;508(1):92-5. doi: 10.1016/j.gene.2012.07.023. Epub 2012 Jul 25. PMID: 22842191.
- Picinelli C, Lintas C, Piras IS, Gabriele S, Sacco R, Brogna C, Persico AM. Recurrent 15q11.2 BP1-BP2 microdeletions and microduplications in the etiology of neurodevelopmental disorders. Am J Med Genet B Neuropsychiatr Genet. 2016 Dec;171(8):1088-1098. doi: 10.1002/ajmg.b.32480. Epub 2016 Aug 26. PMID: 27566550.
- Benítez-Burraco A, Barcos-Martínez M, Espejo-Portero I, Jiménez-Romero S. Variable Penetrance of the 15q11.2 BP1-BP2 Microduplication in a Family with Cognitive and Language Impairment. Mol Syndromol. 2017 May;8(3):139-147. doi: 10.1159/000468192. Epub 2017 Apr 14. PMID: 28588435; PMCID: PMC5448451.
- Sempere Pérez A, Manchón Trives I, Palazón Azorín I, Alcaraz Más L, Pérez Lledó E, Galán Sánchez F. Microdeleción 15q11.2 (BP1-BP2). Un nuevo síndrome con expresividad variable [15Q11.2 (BP1-BP2) microdeletion, a new syndrome with variable expressivity]. An Pediatr (Barc). 2011 Jul;75(1):58-62. Spanish. doi: 10.1016/j.anpedi.2011.01.033. Epub 2011 Mar 17. PMID: 21419731.
- Hashemi B, Bassett A, Chitayat D, Chong K, Feldman M, Flanagan J, Goobie S, Kawamura A, Lowther C, Prasad C, Siu V, So J, Tung S, Speevak M, Stavropoulos DJ, Carter MT. Deletion of 15q11.2(BP1-BP2) region: further evidence for lack of phenotypic specificity in a pediatric population. Am J Med Genet A. 2015 Sep;167A(9):2098-102. doi: 10.1002/ajmg.a.37134. Epub 2015 May 6. PMID: 25946043.
Content Alerts
SignUp to our
Content alerts.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.